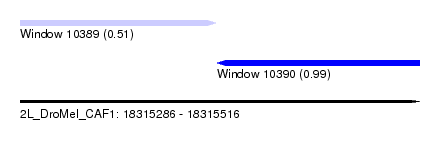
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,315,286 – 18,315,516 |
| Length | 230 |
| Max. P | 0.994148 |

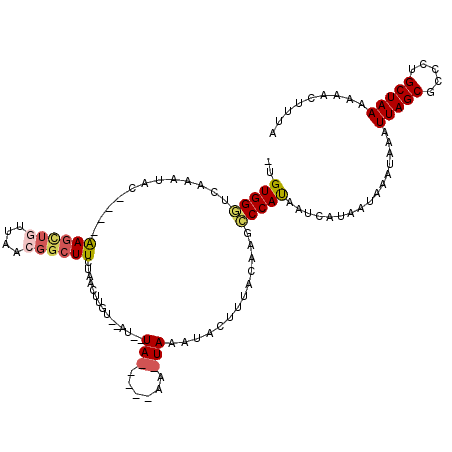
| Location | 18,315,286 – 18,315,399 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.82 |
| Mean single sequence MFE | -19.75 |
| Consensus MFE | -6.94 |
| Energy contribution | -7.92 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.35 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18315286 113 + 22407834 -UGUGGGUCAAAUAC----GACCUAUUAACGGCUUCUAACUUGUAGAUAUUA--GGUAAAUAAAUACUGUACAAACCCAUACUCAUAAUAUUUAAAUUAGCACCAUGCUAAGAAACUUUA -.(((((((......----)))))))....((.((((......(((((((((--((((......))))(((........)))...)))))))))...((((.....)))))))).))... ( -19.60) >DroSec_CAF1 159058 98 + 1 -UGUGGGUCAAAUACAUACAAGCUGUUUACGGCUUAUAACUUGU---------------------ACUUUACAAUCCCAUAAUCAUAAUAAAUAAAUUAGCGCCCUGCUAAAAAACUUUA -((((((............((((((....)))))).....((((---------------------(...))))).))))))...............(((((.....)))))......... ( -18.50) >DroSim_CAF1 167578 92 + 1 -UGUGGGUCAAAUACAUACAAGCUGU------CUUAUAACUUGU---------------------ACUUUACAAUCCCAUAAUCAUAAUAAAUAAAUUAGCGCCAUGCUAAAAAACUUUA -((((((.........((((((.(((------...))).)))))---------------------).........))))))...............(((((.....)))))......... ( -15.77) >DroEre_CAF1 160654 105 + 1 -UGUGGUUCAAA-------AAGUUGUUACCGGCUUCUGGUAUGUUUAUUCUACAACAAAGUAAAUAAUGAAUUAGGCCAC---AGUCGUAAAUAAAUUAGCACCCUGCUAACAAA----G -((((((..((.-------..(((((((((((...))))).((((........)))).....))))))...))..)))))---)............(((((.....)))))....----. ( -20.80) >DroYak_CAF1 170557 116 + 1 AUGUGGGUCAAAUAC----AAGUUGUUAGCGGCUUCUAAUAGGCUUAUUCUACAACGAAGUAAAUAAUUUACUAGGCCAUUUCCAUAAUAAAUAAAUUAGCGCCCUGCUAACAAACUUAG ...............----((((((((((((((..(((((.(((((((((......)))((((.....))))))))))(((.....)))......))))).)))..)))))).))))).. ( -24.10) >consensus _UGUGGGUCAAAUAC____AAGCUGUUAACGGCUUCUAACUUGU__AU__UA_____AA_UAAAUACUUUACAAGCCCAUAAUCAUAAUAAAUAAAUUAGCGCCCUGCUAAAAAACUUUA ..(((((............((((((....))))))...............(((......))).............)))))................(((((.....)))))......... ( -6.94 = -7.92 + 0.98)


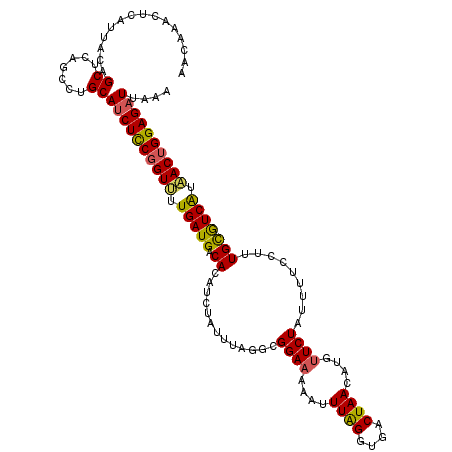
| Location | 18,315,399 – 18,315,516 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -31.34 |
| Consensus MFE | -24.63 |
| Energy contribution | -24.39 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18315399 117 - 22407834 AACAAACUCAUUAGAGCUUAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGUGGAAAAAUUUAGGUGACUAACGUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGUUCAAA .............((((.......))..((((((((.(((((.(((((((....)))))(((....((((....))))....)))..........))))))).)))))))).))... ( -29.60) >DroSec_CAF1 159156 117 - 1 AACAAACUCAUUACAGCUAAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGCGGAAAAUUUUAGGUGACUAAUAUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGAUUAAA ...............((.......))((((((((((.(((((.((..(((....))).(((((((.((((....))))........)))))))..))))))).)))))))))).... ( -30.80) >DroSim_CAF1 167670 117 - 1 AACAAACUCAUUACAGCUUAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGCGGAAAAUUUUGGGUGACUAACAUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGAUUAAA ...............((.......))((((((((((.(((((.((..(((....))).(((((((.((((....))))........)))))))..))))))).)))))))))).... ( -29.80) >DroEre_CAF1 160759 112 - 1 AAAAAACUCAUUACAGCUCAGCCUGCAUCUUCCGUUUUGAUGACACAUCUAUUUAUUGGGAAUAACUUAGGUGACUAA-----UCUACUUUUCUUUGUGUCAUCACUGGAGAUCGAA .......((......((.......))((((.(((...((((((((((((((.....))))......((((....))))-----............)))))))))).))))))).)). ( -25.70) >DroYak_CAF1 170673 117 - 1 AAUAAAUUCAUUACAGCUCAGGCUGCAUCUCCGGUGUUGAUGACACAUAUUUUUAGGUGGAAAAAUUUAGGUGACUAACGUGUUCUAUUUUUCUGUGCAUCGUCACUGGAGAUUCAA .............((((....)))).((((((((((.(((((.((((.......((((((((....((((....))))....))))))))...))))))))).)))))))))).... ( -40.80) >consensus AACAAACUCAUUACAGCUCAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGCGGAAAAAUUUAGGUGACUAACAUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGAUUAAA ...............((.......))((((((((((.(((((.((.............((((....((((....))))....)))).........))))))).)))))))))).... (-24.63 = -24.39 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:57 2006