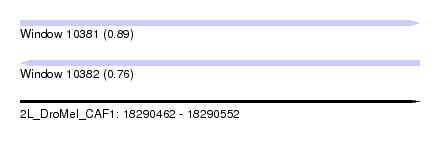
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,290,462 – 18,290,552 |
| Length | 90 |
| Max. P | 0.889484 |

| Location | 18,290,462 – 18,290,552 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.52 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -12.89 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
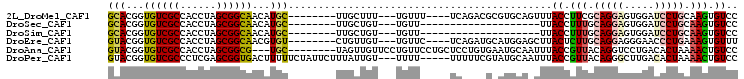
>2L_DroMel_CAF1 18290462 90 + 22407834 GGACACUUGCAGGAUCCACUCCUGCGAAGGUAAACUGCACGCGUCUGA----AAACA---AAAGCAA--------GCAUGUUGCCGCUAGGUGGCGACACCGUGC ...(((((((((((.....)))))))).(((....(((..((...((.----...))---...))..--------))).((((((((...)))))))))))))). ( -31.50) >DroSec_CAF1 134235 74 + 1 GGACACUUGCAGGAUCCACUCCUGCAAAGGUAA--------------------AACA---ACAGCAA--------GCAUGUUGCCGCUAGGUGGCGACACCGUGC ..((..((((((((.....))))))))..))..--------------------....---...(((.--------...(((((((((...)))))))))...))) ( -27.50) >DroSim_CAF1 141250 74 + 1 GGACACUUGCAGGAUCCACUCCUGCAAAGGUAA--------------------AACA---ACAGCAA--------GCAUGUUGCCGCUAGGUGGCGACACCGUGC ..((..((((((((.....))))))))..))..--------------------....---...(((.--------...(((((((((...)))))))))...))) ( -27.50) >DroEre_CAF1 135442 90 + 1 AAACACUUUCAGGGUUCCCUCCUGCAAGAGUAAGCUCCAUGCAUCUGA----GAACA---ACAACAG--------ACACGUUGCCGCUAGGUGGCGACACCGUAC ...........((((((.(...((((.(((....)))..))))...).----)))).---.......--------....((((((((...)))))))).)).... ( -24.90) >DroAna_CAF1 124214 94 + 1 GGACAGUUUUAGUGUCAGGACCUGUAACGGUAAAUUGCAUUCACAGGAGCAGGAACAGGAACAACUA--------GCA---CGCCGCUAGGUGGCGACACCGUAC ...........(((((.(..(((((..(.((...(((......)))..)).)..)))))..).....--------((.---((((....)))))))))))..... ( -25.80) >DroPer_CAF1 156951 97 + 1 GGACAGUUUUAGUGUCAAGCCCUGUAACGGUAAAUUGCAUACGAAAAA-----AAAA---ACAAUAAAGAAUAGAAAAAGUCACCGCUCGAGGGCGACACCGUAC ...........(((((..(((((....((((...(((....)))....-----....---.............((.....))))))....))))))))))..... ( -17.80) >consensus GGACACUUGCAGGAUCCACUCCUGCAAAGGUAAA_UGCAU_C____GA_____AACA___ACAACAA________GCAUGUUGCCGCUAGGUGGCGACACCGUAC ((((.....(((((.....))))).....))................................................((((((((...)))))))).)).... (-12.89 = -13.83 + 0.95)

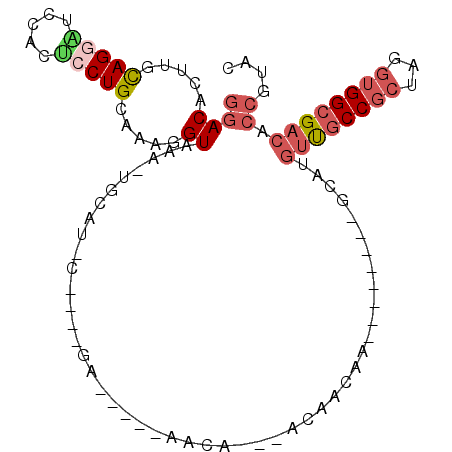
| Location | 18,290,462 – 18,290,552 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.52 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -11.57 |
| Energy contribution | -11.47 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18290462 90 - 22407834 GCACGGUGUCGCCACCUAGCGGCAACAUGC--------UUGCUUU---UGUUU----UCAGACGCGUGCAGUUUACCUUCGCAGGAGUGGAUCCUGCAAGUGUCC ((((((((((((......)))))(((.(((--------.(((.((---((...----.)))).))).)))))).)))...((((((.....))))))..)))).. ( -33.70) >DroSec_CAF1 134235 74 - 1 GCACGGUGUCGCCACCUAGCGGCAACAUGC--------UUGCUGU---UGUU--------------------UUACCUUUGCAGGAGUGGAUCCUGCAAGUGUCC ((((((((....))))(((((((((.....--------)))))))---))..--------------------......((((((((.....)))))))))))).. ( -28.90) >DroSim_CAF1 141250 74 - 1 GCACGGUGUCGCCACCUAGCGGCAACAUGC--------UUGCUGU---UGUU--------------------UUACCUUUGCAGGAGUGGAUCCUGCAAGUGUCC ((((((((....))))(((((((((.....--------)))))))---))..--------------------......((((((((.....)))))))))))).. ( -28.90) >DroEre_CAF1 135442 90 - 1 GUACGGUGUCGCCACCUAGCGGCAACGUGU--------CUGUUGU---UGUUC----UCAGAUGCAUGGAGCUUACUCUUGCAGGAGGGAACCCUGAAAGUGUUU .((((.((((((......)))))).)))).--------.......---.((((----((...((((.((((....))))))))...))))))............. ( -27.10) >DroAna_CAF1 124214 94 - 1 GUACGGUGUCGCCACCUAGCGGCG---UGC--------UAGUUGUUCCUGUUCCUGCUCCUGUGAAUGCAAUUUACCGUUACAGGUCCUGACACUAAAACUGUCC ....(((((((..((((.(..(((---.(.--------.((((((....(((((.......).))))))))))..))))..)))))..))))))).......... ( -23.80) >DroPer_CAF1 156951 97 - 1 GUACGGUGUCGCCCUCGAGCGGUGACUUUUUCUAUUCUUUAUUGU---UUUU-----UUUUUCGUAUGCAAUUUACCGUUACAGGGCUUGACACUAAAACUGUCC ....(((((((((((..((((((((...............(((((---....-----..........)))))))))))))..)))))..)))))).......... ( -25.60) >consensus GCACGGUGUCGCCACCUAGCGGCAACAUGC________UUGCUGU___UGUU_____UC____G_AUGCA_UUUACCUUUGCAGGAGUGGACCCUGAAAGUGUCC (((...((((((......))))))...)))............................................((.(((.(((((.....))))).))).)).. (-11.57 = -11.47 + -0.11)
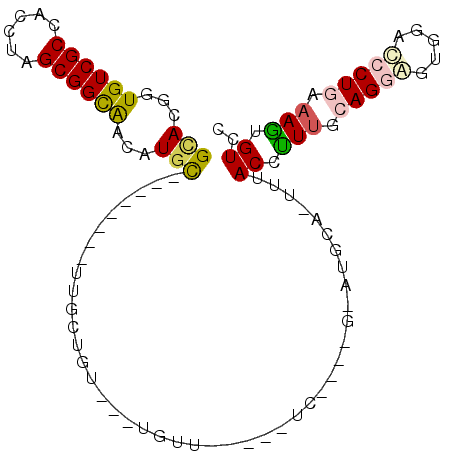
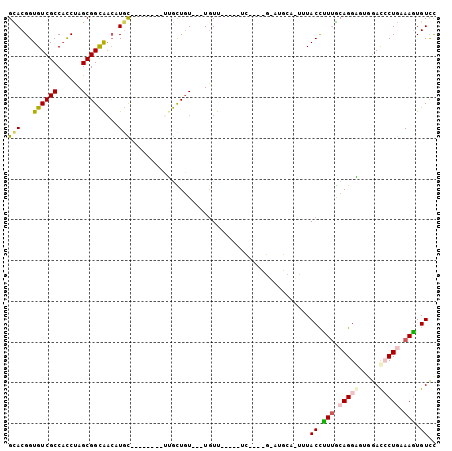
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:49 2006