| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,280,935 – 18,281,055 |
| Length | 120 |
| Max. P | 0.660452 |

| Location | 18,280,935 – 18,281,055 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -45.32 |
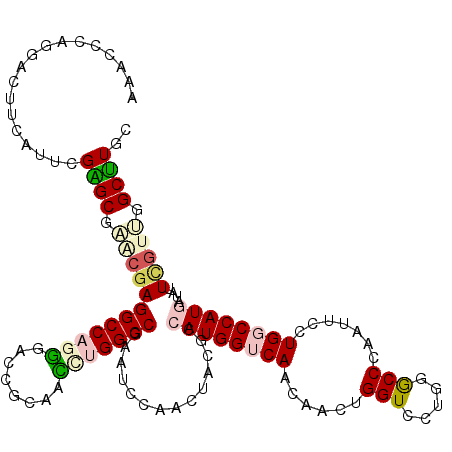
| Consensus MFE | -30.95 |
| Energy contribution | -31.62 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
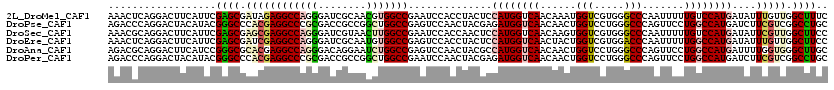
>2L_DroMel_CAF1 18280935 120 - 22407834 AAACUCAGGACUUCAUUCGAGCGAUAGAGGCCAGGGAUCGCAACGUGGCCGAAUCCACCUACUCCAUGGUCAACAAAUGGUCGUGGGCCCAAUUUUUGUCCAUGAUAUUUGUUGGCUUUC .......(((.(((..((........))(((((.(........).)))))))))))...........((((((((((((.((((((((.........))))))))))))))))))))... ( -40.60) >DroPse_CAF1 144444 120 - 1 AGACCCAGGACUACAUACGGGCCCACGAGGCCCGCGACCGCCGGCUGGCCGAGUCCAACUACGAGAUGGUCAACAACUGGUCCUGGGCCCAGUUCCUGGCCAUGAUCUUCGUCGGCCUGC ...((((((((((....((((((.....)))))).(((((((....)))((((.....)).))....))))......))))))))))..........(((((((.....))).))))... ( -50.10) >DroSec_CAF1 124406 120 - 1 AAACGCAGGACUUCAUUCGAGCGAGCGAGGCCAGGGAUCGUAACUUGGCCGAAUCCACCAACUCCAUGGUCAACAAGUGGUCGUGGGCCCAAUUUUUGUCCAUGAUAUUCGUUGGCUUCC .......(((.(((..(((......)))(((((((........)))))))))))))...........(((((((.((((.((((((((.........)))))))))))).)))))))... ( -39.90) >DroEre_CAF1 125781 120 - 1 AAACUCAGGACUUCAUUCGAGCGAUCGAGGCCAGGGAUCGCAAUGUGGCCGAGUCCACCUACUCCAUGGUCAACUACUGGUCGUGGACCCAAUUUUUGGCCAUGAUAUUUGUUGGCUUCC .......((((((...((((....))))(((((.(........).)))))))))))...........(((((((.....(((((((.((........)))))))))....)))))))... ( -40.80) >DroAna_CAF1 116000 120 - 1 AGACGCAGGACUUCAUCCGGGCGCACGAGGCCAGGGACAGGAAUCUGGCCGAGUCCAACUACGCCAUGGUCAACAACUGGUCCUGGGCCCAGUUCCUGGCCAUGAUUUUGGUGGGCUUGC .(.(((.(((.....)))..))))....(((((((........)))))))(((((((.(((...((((((((..((((((.(....).))))))..))))))))....)))))))))).. ( -51.00) >DroPer_CAF1 146602 120 - 1 AGACCCAGGACUACAUACGGGCCCACGAGGCCCGCGACCGCCGGCUGGCCGAAUCCAACUACGAGAUGGUCAACAACUGGUCCUGGGCCCAGUUCCUGGCCAUGAUCUUCGUCGGCCUGC ...((((((((((....((((((.....)))))).((((..(((....))).((((......).)))))))......))))))))))..........(((((((.....))).))))... ( -49.50) >consensus AAACCCAGGACUUCAUUCGAGCGAACGAGGCCAGGGACCGCAACCUGGCCGAAUCCAACUACGCCAUGGUCAACAACUGGUCCUGGGCCCAAUUCCUGGCCAUGAUAUUCGUUGGCUUGC ..................((((.((((((((((((........)))))))..............((((((((......(((.....))).......))))))))....))))).)))).. (-30.95 = -31.62 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:46 2006