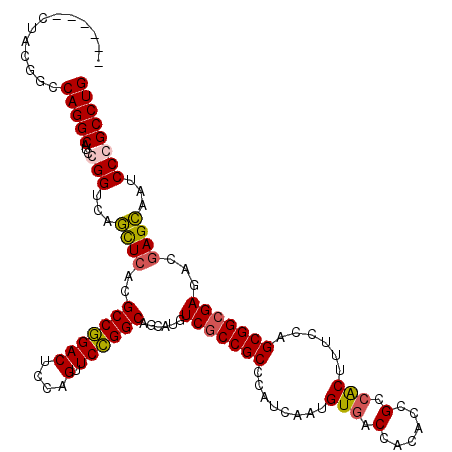
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,951,245 – 1,951,359 |
| Length | 114 |
| Max. P | 0.586307 |

| Location | 1,951,245 – 1,951,359 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -52.50 |
| Consensus MFE | -42.66 |
| Energy contribution | -42.98 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1951245 114 + 22407834 CAGGCGGGAUUACUCGUCUCGCCGCUGGAAAGUGGCGGUGUGGUGACAUUGAUGGGCGGCGACAUGCUGCCGGAACUGGAGUCCGGCGUGAGUUGACCCGAUGCCUGGCCGUAA------ ((((((((.(..((((((((((((((....)))))))).(((....))).))))))..)((((.(((.((((((.(....)))))))))).)))).)))...))))).......------ ( -51.60) >DroEre_CAF1 2894 114 + 1 CAGGCGGGAUUGCUCGUCUCGCCGCUGGAGAGUGGCGGUGUGGUCACAUUGAUGGGCGGCGACAUGCUGCCGGAACUGGAGUCCGGCGUGAGCUGACCCGAGGCCUGGCCGUAG------ ..((((((....))))))((((((((.(.(((((((......))))).))..).))))))))..(((.(((((..((((.(((.(((....)))))))).))..))))).))).------ ( -53.80) >DroYak_CAF1 466 114 + 1 CAGGCGGGAUUGCUCGUCUCGCCGCUGGAUAGUGGCGGUGUGGUCACAUUGAUGGGCGGCGACAUGCUGCCGGAACUGGAGUCCGGCGUGAGCUGACCGGAUGCCUGACCGUAG------ .(((((((....)))))))(((((((....)))))))....(((((........((((((.....))))))((..((((....((((....)))).))))...)))))))....------ ( -50.40) >DroAna_CAF1 23048 120 + 1 CAGGCGGGAUUGCUGGUUUCUCCGCUGGAAAGCGGCGGCGUGGUCACAUUUAUGGGCGGUGACAUGCUGCCAGAACUGGAGUCUGGCGUUAACUGCCCGGAAGCCUGUCCGUGGGGAUAG ..(((((...(((..(...(((((((....)))(((((((((.((((...........)))))))))))))......)))).)..)))....)))))(....).((((((....)))))) ( -54.20) >consensus CAGGCGGGAUUGCUCGUCUCGCCGCUGGAAAGUGGCGGUGUGGUCACAUUGAUGGGCGGCGACAUGCUGCCGGAACUGGAGUCCGGCGUGAGCUGACCCGAUGCCUGGCCGUAG______ ((((((((...(((((((((((((((....)))))))).((.(((.((....))))).))))).....((((((.(....)))))))..))))...)))...)))))............. (-42.66 = -42.98 + 0.31)

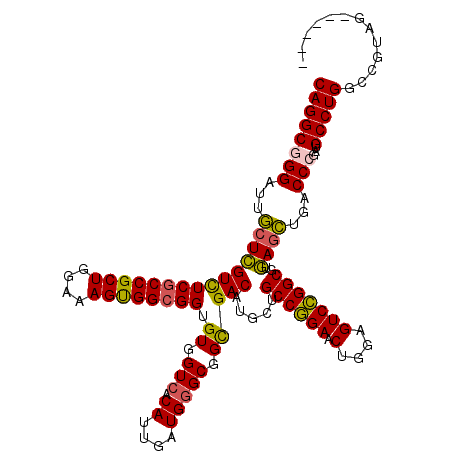
| Location | 1,951,245 – 1,951,359 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -36.81 |
| Energy contribution | -36.94 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1951245 114 - 22407834 ------UUACGGCCAGGCAUCGGGUCAACUCACGCCGGACUCCAGUUCCGGCAGCAUGUCGCCGCCCAUCAAUGUCACCACACCGCCACUUUCCAGCGGCGAGACGAGUAAUCCCGCCUG ------.......(((((...(((...((((..(((((((....).))))))....(.(((((((.......((.(........).)).......))))))).).))))...)))))))) ( -40.04) >DroEre_CAF1 2894 114 - 1 ------CUACGGCCAGGCCUCGGGUCAGCUCACGCCGGACUCCAGUUCCGGCAGCAUGUCGCCGCCCAUCAAUGUGACCACACCGCCACUCUCCAGCGGCGAGACGAGCAAUCCCGCCUG ------.......(((((...(((...((((..(((((((....).))))))....(.(((((((........(((.(......).)))......))))))).).))))...)))))))) ( -44.14) >DroYak_CAF1 466 114 - 1 ------CUACGGUCAGGCAUCCGGUCAGCUCACGCCGGACUCCAGUUCCGGCAGCAUGUCGCCGCCCAUCAAUGUGACCACACCGCCACUAUCCAGCGGCGAGACGAGCAAUCCCGCCUG ------.......(((((....((...((((..(((((((....).))))))....(.(((((((........(((.(......).)))......))))))).).))))...)).))))) ( -40.54) >DroAna_CAF1 23048 120 - 1 CUAUCCCCACGGACAGGCUUCCGGGCAGUUAACGCCAGACUCCAGUUCUGGCAGCAUGUCACCGCCCAUAAAUGUGACCACGCCGCCGCUUUCCAGCGGAGAAACCAGCAAUCCCGCCUG ...(((....)))(((((.....(((.......(((((((....).)))))).....(((((...........)))))...))).(((((....)))))................))))) ( -36.10) >consensus ______CUACGGCCAGGCAUCCGGUCAGCUCACGCCGGACUCCAGUUCCGGCAGCAUGUCGCCGCCCAUCAAUGUGACCACACCGCCACUUUCCAGCGGCGAGACGAGCAAUCCCGCCUG .............(((((...(((...((((..(((((((....).))))))......(((((((........(((.(......).)))......)))))))...))))...)))))))) (-36.81 = -36.94 + 0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:55 2006