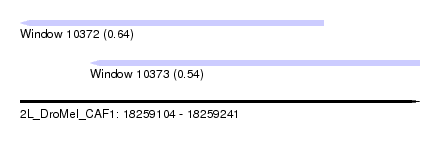
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,259,104 – 18,259,241 |
| Length | 137 |
| Max. P | 0.643989 |

| Location | 18,259,104 – 18,259,208 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -16.32 |
| Energy contribution | -17.76 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
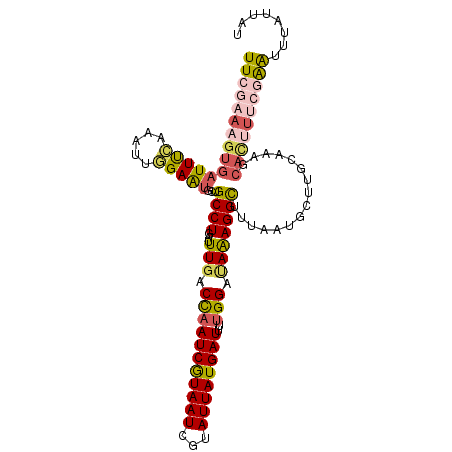
>2L_DroMel_CAF1 18259104 104 - 22407834 UUGGAAAGUGAUUCCAAAUUGGAACGCUGCCUGAUUUGACCAAUCGUAAUAGUAUUAUGAUUUUGGGUAAAGGCUUUAAUGCUUGCAAAGCAUUUUCGAACUCA---- ((((((((((.((((.....))))))))((((......((((((((((((...)))))))))...)))..))))....(((((.....))))))))))).....---- ( -25.80) >DroSec_CAF1 102486 108 - 1 UUCGAAAGUGAUUUCAAAUUGGAAUGCUGCCUGAUUUGACCAAUCGUAAUCGUAUUAUGAUUUUGGAUAAAGGCUUUAAUGCUUGCAAAGCACUUUCGAAUUUAUUAU ((((((((((...((((((..(........)..))))))(((((((((((...))))))))..)))....((((......))))......))))))))))........ ( -29.50) >DroSim_CAF1 108251 108 - 1 UUCGAAAGUGAUUUCAAAUUGGAAUGCUGCCUGAUUUGACCAAUCGUAAUCGUAUUAUGAUUUUGGAUAAAGGCUUUAAUGCUUGCAAAGCACUUUCGAAUUUAUUAU ((((((((((...((((((..(........)..))))))(((((((((((...))))))))..)))....((((......))))......))))))))))........ ( -29.50) >DroEre_CAF1 103728 108 - 1 UUCAAAAGUGAUUUCAAAUUUGAAUGCUGCCUGAGUUGACUAAUCGUAAUCGUAUUAUGAUUUUGGACAGAGGUUUUAGUACUUACAAAGCACCUAGGAAUUUAUUUU ((((((..((....))..))))))((((((((..(((.(..(((((((((...))))))))).).)))..))))....((....))..))))................ ( -18.90) >DroYak_CAF1 107693 95 - 1 UUCGAAAGUGAUUUGAAAUUCGAGUUCUGCCUGAUUUUUCCCAUCAUAAUCGUAUUAUGAUUUUGGACAAAGGCUUUAAUACU-AC------------GGUUUAUUUU ((((((.....))))))..((((((...((((......(((.((((((((...))))))))...)))...))))......)))-.)------------))........ ( -16.50) >consensus UUCGAAAGUGAUUUCAAAUUGGAAUGCUGCCUGAUUUGACCAAUCGUAAUCGUAUUAUGAUUUUGGAUAAAGGCUUUAAUGCUUGCAAAGCACUUUCGAAUUUAUUAU (((((((((((((((.....)))))...((((...(((.(((((((((((...))))))))..))).)))))))................))))))))))........ (-16.32 = -17.76 + 1.44)

| Location | 18,259,128 – 18,259,241 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.36 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
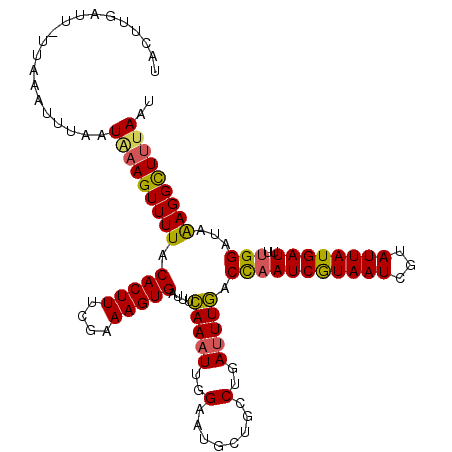
>2L_DroMel_CAF1 18259128 113 - 22407834 UAUUUGAAU-UUAAAUUUAAUAAAGUUUUACACUUUGGAAAGUGAUUCCAAAUUGGAACGCUGCCUGAUUUGACCAAUCGUAAUAGUAUUAUGAUUUUGGGUAAAGGCUUUAAU ...((((((-....))))))(((((((((.((..(..(..((((.((((.....))))))))..)..)..)).(((((((((((...))))))))..)))...))))))))).. ( -25.40) >DroSec_CAF1 102514 113 - 1 UACUUGAUU-UUAAAUUUAAUAAAGUUUUACACUUUCGAAAGUGAUUUCAAAUUGGAAUGCUGCCUGAUUUGACCAAUCGUAAUCGUAUUAUGAUUUUGGAUAAAGGCUUUAAU .........-..........(((((((((.(((((....)))))...((((((..(........)..))))))(((((((((((...))))))))..)))...))))))))).. ( -23.00) >DroSim_CAF1 108279 113 - 1 UACUUGAUU-UUAAAUUUAAUAAAGUUUUACACUUUCGAAAGUGAUUUCAAAUUGGAAUGCUGCCUGAUUUGACCAAUCGUAAUCGUAUUAUGAUUUUGGAUAAAGGCUUUAAU .........-..........(((((((((.(((((....)))))...((((((..(........)..))))))(((((((((((...))))))))..)))...))))))))).. ( -23.00) >DroEre_CAF1 103756 99 - 1 --------------A-UUAAUGUAGUUUUACACUUUCAAAAGUGAUUUCAAAUUUGAAUGCUGCCUGAGUUGACUAAUCGUAAUCGUAUUAUGAUUUUGGACAGAGGUUUUAGU --------------.-....((((....))))..((((((..((....))..)))))).(((((((..(((.(..(((((((((...))))))))).).)))..))))...))) ( -16.70) >DroYak_CAF1 107708 114 - 1 UACUUGAUUUUUAAAUUAAAUCAAGUUUUACACUUUCGAAAGUGAUUUGAAAUUCGAGUUCUGCCUGAUUUUUCCCAUCAUAAUCGUAUUAUGAUUUUGGACAAAGGCUUUAAU .((((((.((((((((((..((((((.....))))..))...)))))))))).))))))...((((......(((.((((((((...))))))))...)))...))))...... ( -25.40) >consensus UACUUGAUU_UUAAAUUUAAUAAAGUUUUACACUUUCGAAAGUGAUUUCAAAUUGGAAUGCUGCCUGAUUUGACCAAUCGUAAUCGUAUUAUGAUUUUGGAUAAAGGCUUUAAU ....................(((((((((.(((((....)))))....(((((..(........)..))))).(((((((((((...))))))))..)))...))))))))).. (-16.90 = -16.62 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:40 2006