| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,256,684 – 18,256,780 |
| Length | 96 |
| Max. P | 0.865653 |

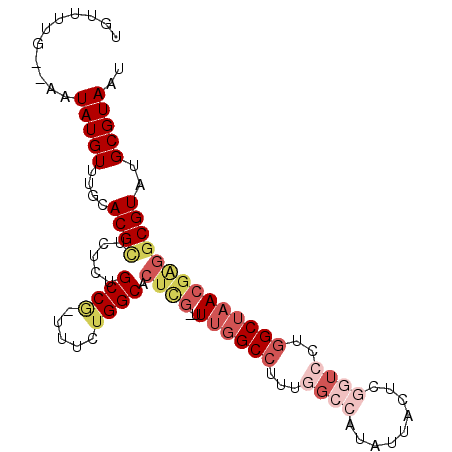
| Location | 18,256,684 – 18,256,780 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -15.36 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18256684 96 + 22407834 AUUACGCAUACGCCUCGUUAGCCAGGACCGAGUAAUAAGGCCAAAGGCCAAA-CGAGUGCCAGAAA-CGGCAGAGAGCGUGCAAACAUAUU--CAAAACA ..(((((......((((((.(((.((.((.........))))...)))..))-))))((((.....-.))))....)))))..........--....... ( -25.90) >DroVir_CAF1 160024 91 + 1 AUUACGCAUACGCAGCGUAUGCA-GCAGAAAAUUGUUUGGCUGCGGGCCAAACCAAAUGCCA--AA-UGGCGAAA--CGUG-AAACAUAUU--UUCAAUA .....(((((((...))))))).-...(((((((((((.((...((......))...((((.--..-.))))...--.)).-))))).)))--))).... ( -27.10) >DroPse_CAF1 115952 91 + 1 AUUACGCAUACGCCCGGUUAGCCAG---CGAGUAAUUUGGCUAAAGGCCAAA--AAGUGCCCAAAACCGGCA---AACGUA-AAACAUAUUUUGAAAACA .(((((.....(((...((((((((---........)))))))).)))....--...((((.......))))---..))))-)................. ( -21.80) >DroSim_CAF1 105820 96 + 1 AUUACGCAUACGCCUCGUUAGCCAGGACCGAGUAAUAUGGCCAAAGGCCAAA-CGAGUGCCAGAAA-CGGCAGAGAGCGUGCAAACAUAUU--CAAAACA ..(((((......((((((.(((.((.((((....).)))))...)))..))-))))((((.....-.))))....)))))..........--....... ( -26.00) >DroEre_CAF1 101265 96 + 1 AUUACGCAUACGCCUCGUUAGCCAGGACGGAAUAAUAUGGCCAAAGGCCAAC-CGAGUGCCAGAAA-CGGCGGAGAGCGUGCAAACAUAUU--CAAAACA ..(((((...(((((((((......))))).......(((((...)))))..-.............-.))))....)))))..........--....... ( -23.60) >DroYak_CAF1 105163 96 + 1 AUUACGCAUACGCCUCGUUAGCCAGGACCGAGUAAUAUGGCCAAAGGCCAAA-CGAGUGCCAGAAA-CGGCAGAGAGCGUGCAAACAUAUU--CAAAACA ..(((((......((((((.(((.((.((((....).)))))...)))..))-))))((((.....-.))))....)))))..........--....... ( -26.00) >consensus AUUACGCAUACGCCUCGUUAGCCAGGACCGAGUAAUAUGGCCAAAGGCCAAA_CGAGUGCCAGAAA_CGGCAGAGAGCGUGCAAACAUAUU__CAAAACA ........((((((((.....................(((((...))))).......((((.......))))))).)))))................... (-15.36 = -16.28 + 0.92)

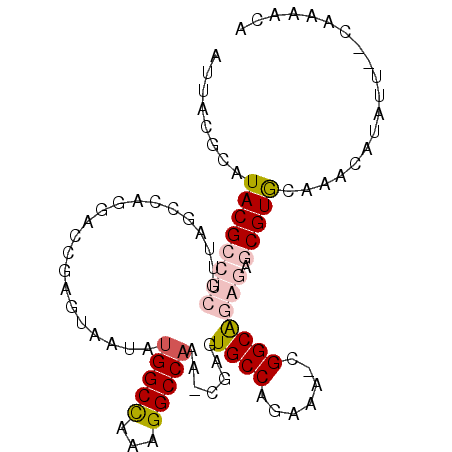
| Location | 18,256,684 – 18,256,780 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.88 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -18.29 |
| Energy contribution | -20.52 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18256684 96 - 22407834 UGUUUUG--AAUAUGUUUGCACGCUCUCUGCCG-UUUCUGGCACUCG-UUUGGCCUUUGGCCUUAUUACUCGGUCCUGGCUAACGAGGCGUAUGCGUAAU .......--..(((((....((((....(((((-....)))))((((-((.((((...((((.........))))..))))))))))))))..))))).. ( -31.60) >DroVir_CAF1 160024 91 - 1 UAUUGAA--AAUAUGUUU-CACG--UUUCGCCA-UU--UGGCAUUUGGUUUGGCCCGCAGCCAAACAAUUUUCUGC-UGCAUACGCUGCGUAUGCGUAAU ....(((--(((......-....--....(((.-..--.))).....(((((((.....))))))).))))))(((-.(((((((...)))))))))).. ( -27.40) >DroPse_CAF1 115952 91 - 1 UGUUUUCAAAAUAUGUUU-UACGUU---UGCCGGUUUUGGGCACUU--UUUGGCCUUUAGCCAAAUUACUCG---CUGGCUAACCGGGCGUAUGCGUAAU ...........(((((..-(((((.---..((((((.(.(((....--((((((.....))))))......)---)).)..))))))))))).))))).. ( -26.90) >DroSim_CAF1 105820 96 - 1 UGUUUUG--AAUAUGUUUGCACGCUCUCUGCCG-UUUCUGGCACUCG-UUUGGCCUUUGGCCAUAUUACUCGGUCCUGGCUAACGAGGCGUAUGCGUAAU .......--..(((((....((((....(((((-....)))))((((-((.((((...((((.........))))..))))))))))))))..))))).. ( -31.70) >DroEre_CAF1 101265 96 - 1 UGUUUUG--AAUAUGUUUGCACGCUCUCCGCCG-UUUCUGGCACUCG-GUUGGCCUUUGGCCAUAUUAUUCCGUCCUGGCUAACGAGGCGUAUGCGUAAU .......--..(((((....((((.(((.((((-....)))).....-(((((((...(((...........)))..))))))))))))))..))))).. ( -28.10) >DroYak_CAF1 105163 96 - 1 UGUUUUG--AAUAUGUUUGCACGCUCUCUGCCG-UUUCUGGCACUCG-UUUGGCCUUUGGCCAUAUUACUCGGUCCUGGCUAACGAGGCGUAUGCGUAAU .......--..(((((....((((....(((((-....)))))((((-((.((((...((((.........))))..))))))))))))))..))))).. ( -31.70) >consensus UGUUUUG__AAUAUGUUUGCACGCUCUCUGCCG_UUUCUGGCACUCG_UUUGGCCUUUGGCCAUAUUACUCGGUCCUGGCUAACGAGGCGUAUGCGUAAU ...........(((((....((((.....((((.....)))).((((..((((((...((((.........))))..))))))))))))))..))))).. (-18.29 = -20.52 + 2.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:38 2006