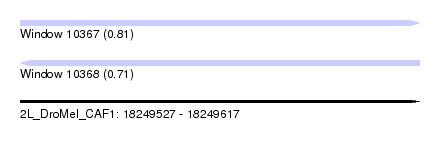
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,249,527 – 18,249,617 |
| Length | 90 |
| Max. P | 0.813295 |

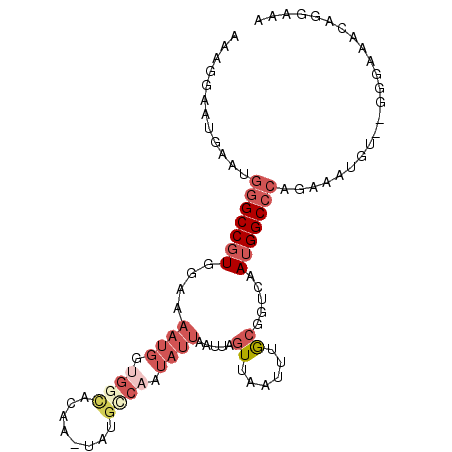
| Location | 18,249,527 – 18,249,617 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -15.67 |
| Consensus MFE | -9.20 |
| Energy contribution | -9.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18249527 90 + 22407834 AUUC-------CC--ACAUUUCUGGGCCAUUGACCGCAAAUUAACUAAUUAAUAUUGACAGAACAGCGCCACCAUUUUCCACGGCCCAUUCUUUCCUUU ....-------..--.......((((((.(((....)))(((((....))))).............................))))))........... ( -11.50) >DroSec_CAF1 92764 97 + 1 UUUCCCUUUACCC--ACAUUUCUGGGCCAUUGACCGCAAAUUAACUAAUUAAUAUUGGCAGAACAGUGCCACCAUUUUCCACGGCCCAUUCAUUCCUUU .............--.......((((((.(((....)))................(((((......)))))...........))))))........... ( -18.20) >DroSim_CAF1 98436 97 + 1 UUUCCCUUUACCC--ACAUUUCUGGGCCAUUGACCGCAAAUUAACUAAUUAAUAUUGGCAGAACAGUACCACCAUUGUCCACGGCCCAUUCAUUCCUUU .............--.......((((((..((.(((...(((((....)))))..)))))(.(((((......))))).)..))))))........... ( -16.90) >DroEre_CAF1 94355 95 + 1 UGUCCAGUUUCCC--ACAUUUCUGGGCCAUUGACCGCAAAUUAACUAAUUAAUAUUGGCCUG-UUCUGCCGCCAUU-UGCACGGCCCAUUCAUUCCUUU .............--.......((((((.......((((((...(((((....)))))...)-)).))).((....-.))..))))))........... ( -17.90) >DroYak_CAF1 98222 95 + 1 UUUCCUGUUUCCC--ACAUUUCUGGGCCAUUGACCGCAAAUUAACUAAUUAAUAUUGGCCUG-UUCUGACACCAUU-UCCACGGCCCGUUCAUUCCUUU .............--........(((((.(((....)))................(((..((-(....))))))..-.....)))))............ ( -13.30) >DroPer_CAF1 108383 84 + 1 GUUCGUGCCCCCCAAAUAUUUCUGAGCCAUUGAAUCUUAAUUAACUAAUUAAAAUUUGCUUA-UU-----------UGGCAGGGC--UUU-UAUCCUUU ......((((.(((((((......(((....((((.((((((....)))))).)))))))))-))-----------)))..))))--...-........ ( -16.20) >consensus UUUCCCGUUACCC__ACAUUUCUGGGCCAUUGACCGCAAAUUAACUAAUUAAUAUUGGCAGA_CAGUGCCACCAUUUUCCACGGCCCAUUCAUUCCUUU ......................((((((.(((....))).....(((((....)))))........................))))))........... ( -9.20 = -9.95 + 0.75)

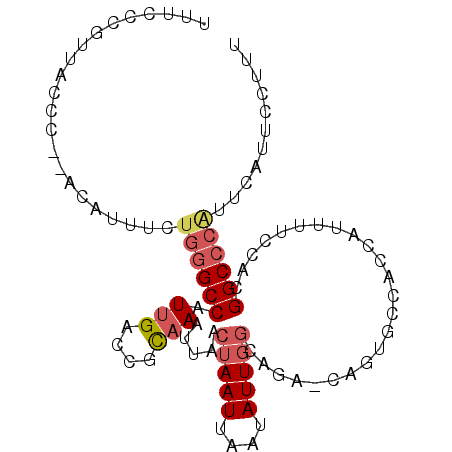

| Location | 18,249,527 – 18,249,617 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -10.45 |
| Energy contribution | -12.45 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18249527 90 - 22407834 AAAGGAAAGAAUGGGCCGUGGAAAAUGGUGGCGCUGUUCUGUCAAUAUUAAUUAGUUAAUUUGCGGUCAAUGGCCCAGAAAUGU--GG-------GAAU ...........((((((((.((.((((.(((((......))))).)))).....((......))..)).)))))))).......--..-------.... ( -19.90) >DroSec_CAF1 92764 97 - 1 AAAGGAAUGAAUGGGCCGUGGAAAAUGGUGGCACUGUUCUGCCAAUAUUAAUUAGUUAAUUUGCGGUCAAUGGCCCAGAAAUGU--GGGUAAAGGGAAA .............(((((..((......(((((......)))))...((((....))))))..)))))....(((((......)--))))......... ( -23.90) >DroSim_CAF1 98436 97 - 1 AAAGGAAUGAAUGGGCCGUGGACAAUGGUGGUACUGUUCUGCCAAUAUUAAUUAGUUAAUUUGCGGUCAAUGGCCCAGAAAUGU--GGGUAAAGGGAAA ...........((((((((.(((..(((..(.......)..)))..........((......)).))).)))))))).......--............. ( -25.20) >DroEre_CAF1 94355 95 - 1 AAAGGAAUGAAUGGGCCGUGCA-AAUGGCGGCAGAA-CAGGCCAAUAUUAAUUAGUUAAUUUGCGGUCAAUGGCCCAGAAAUGU--GGGAAACUGGACA ...........((((((((...-..((((.(((((.-..(((.(((....))).)))..))))).)))))))))))).......--((....))..... ( -26.10) >DroYak_CAF1 98222 95 - 1 AAAGGAAUGAACGGGCCGUGGA-AAUGGUGUCAGAA-CAGGCCAAUAUUAAUUAGUUAAUUUGCGGUCAAUGGCCCAGAAAUGU--GGGAAACAGGAAA ............(((((((...-....((......)-).((((...(((((....)))))....)))).)))))))........--.(....)...... ( -21.10) >DroPer_CAF1 108383 84 - 1 AAAGGAUA-AAA--GCCCUGCCA-----------AA-UAAGCAAAUUUUAAUUAGUUAAUUAAGAUUCAAUGGCUCAGAAAUAUUUGGGGGGCACGAAC ........-...--(((((.(((-----------((-(((((.((((((((((....)))))))))).....)))......))))))))))))...... ( -21.70) >consensus AAAGGAAUGAAUGGGCCGUGGAAAAUGGUGGCACAA_UAUGCCAAUAUUAAUUAGUUAAUUUGCGGUCAAUGGCCCAGAAAUGU__GGGAAACAGGAAA ............(((((((....((((.((((........)))).)))).....((......)).....)))))))....................... (-10.45 = -12.45 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:35 2006