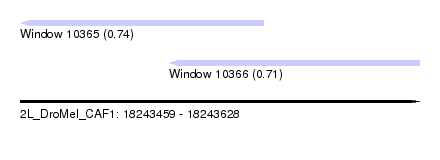
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,243,459 – 18,243,628 |
| Length | 169 |
| Max. P | 0.735935 |

| Location | 18,243,459 – 18,243,562 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -17.13 |
| Energy contribution | -17.02 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
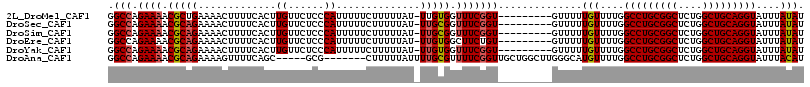
>2L_DroMel_CAF1 18243459 103 - 22407834 UUUUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUAUAUUUAAAUGCGGCUUGCGGUUCGCCAUUGCGCCUAUUGUUAUGCAAUGAAAGUUU--GUUCGCG--------------AAC- ...(((...((((((((((....)))))).(((((((....))))))))))).)))(((((((((((((..........)))))))...(...--...))))--------------)))- ( -33.10) >DroVir_CAF1 142333 105 - 1 ---UUUUCUGGC--------CGAGCUGCAGGUAUUUAUAUUUAAAUGCUGCUUGUGGCACGCCAUUGCGCCUAUUGUUAUGCAAUGAAAGUUU-UGUGUUCGCAUUGUUCUUG---CACG ---.....((((--------.(.((..((((((((((....))))))))...))..)).)))))...............(((((.(((.....-(((....)))...))))))---)).. ( -25.90) >DroPse_CAF1 98218 106 - 1 -----UUUCAGCCUGUG-----GGCUGCAGGUAUUUACAUUUAAAUGCUGCUUGCGGUUCGCCAGUGCGCCUAUUGUUAUGCAAUGAAAGUUUUUGUACGCGCAUUGUUCGCC---ACC- -----.....((..(((-----(((((((((((((((....))))))))...))))))))))((((((((.(((((.....)))))...((......))))))))))...)).---...- ( -33.70) >DroWil_CAF1 123959 108 - 1 GGUUUUUCUGGCC--------GAGCUGCAGGUAUUUAUAUUUAAAUGCUGUUUGUGGUACGCCAUUGCGCCUAUUGUUAUGCAAUGAAACUUU--GUUCUCGCAUUGUUUUCA-GCAUC- (((.....((((.--------(.((..((((((((((....))))))))...))..)).)))))....))).......((((...(((((..(--((....)))..)))))..-)))).- ( -26.80) >DroAna_CAF1 81115 117 - 1 GCAUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUACAUUUAAAUGCGGCCUGUGGUUCGCCAUUGCGCCUAUUGUUAUGCAAUGAAAGUUU--GUCCGCGCAUUGUUCACAAGGACC- ..((((....(((((((((....)))))))))....))))...(((((((((.((((....)))).).)))(((((.....))))).......--......)))))((((....)))).- ( -39.10) >DroPer_CAF1 99608 106 - 1 -----UUUCAGCCUGUG-----GGCUGCAGGUAUUUACAUUUAAAUGCUGCUUGCGGCUCGCCAGUGCGCCUAUUGUUAUGCAAUGAAAGUUUUUGUACGCGCAUUGUUCACC---ACC- -----.........(((-----(((((((((((((((....))))))))...))))))))))((((((((.(((((.....)))))...((......))))))))))......---...- ( -35.30) >consensus ___U_UUUUGGCCUG_G_____GGCUGCAGGUAUUUACAUUUAAAUGCUGCUUGCGGUUCGCCAUUGCGCCUAUUGUUAUGCAAUGAAAGUUU__GUACGCGCAUUGUUCACA___ACC_ ..........((...........((((((((((((((....)))))))...)))))))....(((((((..........)))))))...............))................. (-17.13 = -17.02 + -0.11)

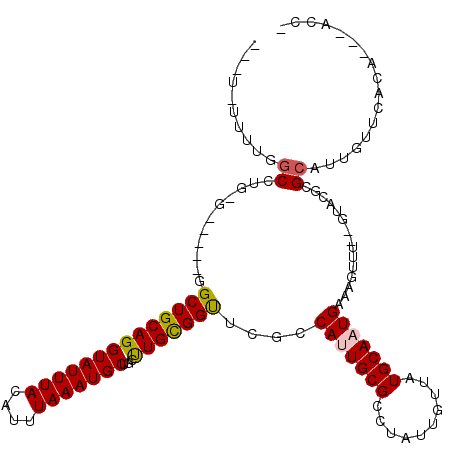
| Location | 18,243,522 – 18,243,628 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.48 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18243522 106 - 22407834 GGCCAGAAAACGCUGAAAACUUUUCACUUGUUCUCCCAUUUUUCUUUUUAU-UUGUGGUUUCGGU---------GUUUUUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUAUAU ((((((((((((((((((.....((((..((..................))-..)))))))))))---------))))))....)))))((((((....))))))........... ( -31.07) >DroSec_CAF1 86744 106 - 1 GGCCAGAAAACGCAGAAAACUUUUCACUUGUUCUCCCAUUUUUCUUUUUAU-UUGCGGUUUCGGU---------GUUUUUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUAUAU .(((.((((.((((((............((......))............)-))))).)))))))---------.....(((....(((((((((....)))))))))....))). ( -28.75) >DroSim_CAF1 90886 106 - 1 GGCCAGAAAACGCAGAAAACUUUUCACUUGUUCUCCCAUUUUUCUUUUUAU-UUGCGGUUUCGGU---------GUUUUUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUAUAU .(((.((((.((((((............((......))............)-))))).)))))))---------.....(((....(((((((((....)))))))))....))). ( -28.75) >DroEre_CAF1 88334 106 - 1 GGCCAGAAAACGCAGAAAACUUUUCACUUGUUCUCCCAUUUUUCUUUUUAU-UUGUGGCUUCUGU---------GUUUUUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUAUAU ((((((((((((((((((((.........)))...((((............-..)))).))))))---------))))))....)))))((((((....))))))........... ( -31.14) >DroYak_CAF1 92212 106 - 1 GGCCAGAAAACGCAGAAAACUUUUCACUUGUUCUCCCAUUUUUCUUUUUAU-UUGUGGUUUCGGU---------GUUUUUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUAUAU (((((((((((((.((((.....((((..((..................))-..)))))))).))---------))))))....)))))((((((....))))))........... ( -27.17) >DroAna_CAF1 81192 104 - 1 GGCCAGAAAACGCAGAAAAGUUUUCAGC-----GCG-------CUUUUUAUUUUGCGUUUUCGGUUGCUGGCUUGGGCAUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUACAU ((((.((((((((((((((((.......-----..)-------))))))....)))))))))))))(((......)))((((....(((((((((....)))))))))....)))) ( -39.90) >consensus GGCCAGAAAACGCAGAAAACUUUUCACUUGUUCUCCCAUUUUUCUUUUUAU_UUGCGGUUUCGGU_________GUUUUUGUUUUGGCCUGCGGCUCUGGCUGCAGGUAUUUAUAU .(((.((((.(((((.............((......))..............))))).)))))))..............(((....(((((((((....)))))))))....))). (-23.70 = -23.90 + 0.20)
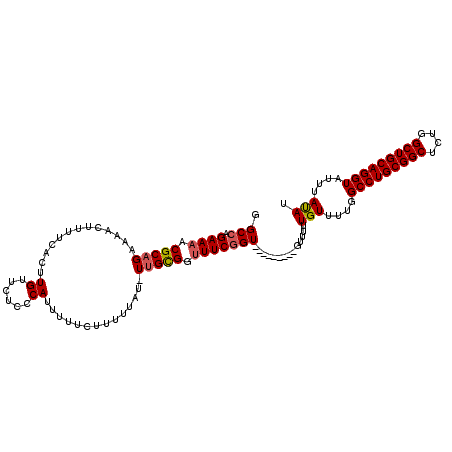
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:34 2006