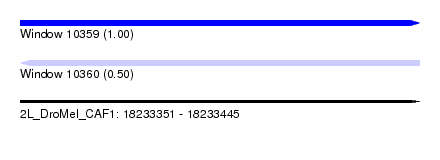
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,233,351 – 18,233,445 |
| Length | 94 |
| Max. P | 0.995442 |

| Location | 18,233,351 – 18,233,445 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -5.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18233351 94 + 22407834 AAGAA-AUUUUCUCUAAUGGCUUUCUUUGGUUGGCCACAACGGAAACAAAAGACCAAAUGCCAAAGCCACAAAAGGAGAGAAAAUUUCAAUUCAA ..(((-(((((((((..(((((((.((((((((....)...(....)....)))))))....))))))).......))))))))))))....... ( -28.60) >DroSec_CAF1 76503 94 + 1 AAGAA-AUUUUCUCUAAUGGCUUUCUUCGGUUGGCCACAACGGAAACAAAAGACCAAAUGCCAAAGCCACAAAAGGAGAGAAAAUUUCAAUUCAA ..(((-(((((((((..(((((((..((.((((....))))(....)....))..))).))))...((......))))))))))))))....... ( -27.80) >DroSim_CAF1 80670 94 + 1 AAGAA-AUUUUCUCUAAUGGCUUUCUUCGGUUGGCCACAACGGAAACAAAAGACCAAAUGCCAAAGCCACAAAAGGAGAGAAAAUUUCAAUUCAA ..(((-(((((((((..(((((((..((.((((....))))(....)....))..))).))))...((......))))))))))))))....... ( -27.80) >DroEre_CAF1 78060 94 + 1 AAGAA-AUUUUCUCUAAUGGCUUUCUUUGGUUGGCCACAAAGGAAACAAAAGACCAAAUGCCAAAGCCGCAAAAGGAGAGAAAAUUUCAAUUCAA ..(((-(((((((((..((((....((((((((....)...(....)....))))))).))))...((......))))))))))))))....... ( -29.20) >DroYak_CAF1 80922 94 + 1 AAGAA-AUUUUCUCUAAUGGCUUUCUUUGGUUGGCCACAAAGGAAACAAAAGACCAAAUGCCAAAGCCACAAAAGGAGAGAAAAUUUCAAUUCAA ..(((-(((((((((..(((((((.((((((((....)...(....)....)))))))....))))))).......))))))))))))....... ( -28.50) >DroAna_CAF1 71107 95 + 1 GGGAAAAUUUCCUUUAAUGGCUUUUGUUGGUCGCCCGCAAUGGAAACAAAAGACCAAAGCCAAAUGCCGCAAAAGGAGAGAAAUUUUCAAUUCAA (..((..((((((((..((((.(((((((((((....)..((....))...))))))...)))).))))..))))))))....))..)....... ( -28.50) >consensus AAGAA_AUUUUCUCUAAUGGCUUUCUUUGGUUGGCCACAACGGAAACAAAAGACCAAAUGCCAAAGCCACAAAAGGAGAGAAAAUUUCAAUUCAA ..(((.(((((((((..(((((((.((((((((....)...(....)....)))))))....))))))).......))))))))))))....... (-21.42 = -21.92 + 0.50)

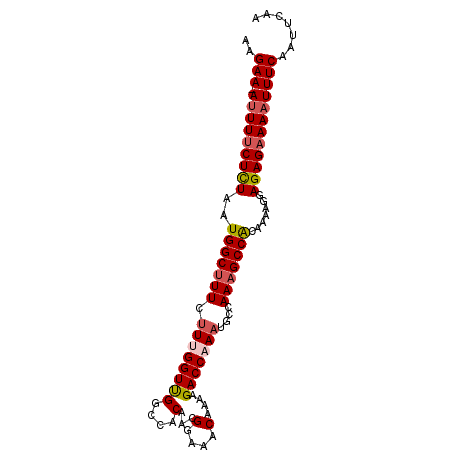
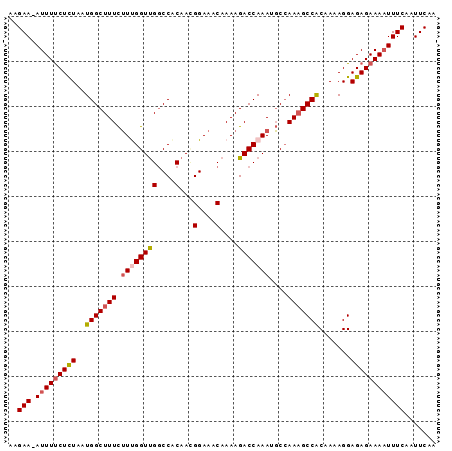
| Location | 18,233,351 – 18,233,445 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.79 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18233351 94 - 22407834 UUGAAUUGAAAUUUUCUCUCCUUUUGUGGCUUUGGCAUUUGGUCUUUUGUUUCCGUUGUGGCCAACCAAAGAAAGCCAUUAGAGAAAAU-UUCUU .......((((((((((((......((((((((((...((((((...............)))))))))....))))))).)))))))))-))).. ( -28.46) >DroSec_CAF1 76503 94 - 1 UUGAAUUGAAAUUUUCUCUCCUUUUGUGGCUUUGGCAUUUGGUCUUUUGUUUCCGUUGUGGCCAACCGAAGAAAGCCAUUAGAGAAAAU-UUCUU .......((((((((((((......(((((((((((.....))).....((((.((((....)))).)))))))))))).)))))))))-))).. ( -28.50) >DroSim_CAF1 80670 94 - 1 UUGAAUUGAAAUUUUCUCUCCUUUUGUGGCUUUGGCAUUUGGUCUUUUGUUUCCGUUGUGGCCAACCGAAGAAAGCCAUUAGAGAAAAU-UUCUU .......((((((((((((......(((((((((((.....))).....((((.((((....)))).)))))))))))).)))))))))-))).. ( -28.50) >DroEre_CAF1 78060 94 - 1 UUGAAUUGAAAUUUUCUCUCCUUUUGCGGCUUUGGCAUUUGGUCUUUUGUUUCCUUUGUGGCCAACCAAAGAAAGCCAUUAGAGAAAAU-UUCUU .......((((((((((((......((..((((((...((((((...............))))))))))))...))....)))))))))-))).. ( -28.26) >DroYak_CAF1 80922 94 - 1 UUGAAUUGAAAUUUUCUCUCCUUUUGUGGCUUUGGCAUUUGGUCUUUUGUUUCCUUUGUGGCCAACCAAAGAAAGCCAUUAGAGAAAAU-UUCUU .......((((((((((((......((((((((((...((((((...............)))))))))....))))))).)))))))))-))).. ( -28.46) >DroAna_CAF1 71107 95 - 1 UUGAAUUGAAAAUUUCUCUCCUUUUGCGGCAUUUGGCUUUGGUCUUUUGUUUCCAUUGCGGGCGACCAACAAAAGCCAUUAAAGGAAAUUUUCCC .......(((((((....((((((...(((.((((((..(((.(....)...)))..))((....))..)))).)))...))))))))))))).. ( -26.70) >consensus UUGAAUUGAAAUUUUCUCUCCUUUUGUGGCUUUGGCAUUUGGUCUUUUGUUUCCGUUGUGGCCAACCAAAGAAAGCCAUUAGAGAAAAU_UUCUU .......((((((((((((......((((((((.....((((((...............))))))......)))))))).))))))))).))).. (-19.22 = -19.94 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:28 2006