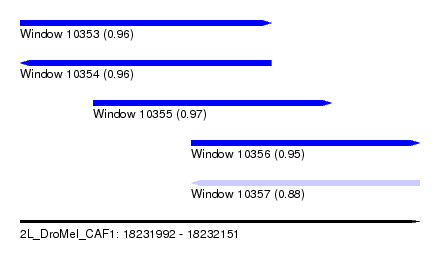
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,231,992 – 18,232,151 |
| Length | 159 |
| Max. P | 0.968695 |

| Location | 18,231,992 – 18,232,092 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -18.57 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
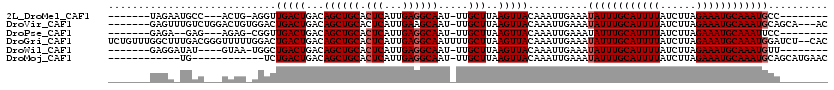
>2L_DroMel_CAF1 18231992 100 + 22407834 -------UAGAAUGCC---ACUG-AGGUUGACUGACAGCUGCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCC-------- -------..((.(((.---.(((-.((....))..)))..))).)).....((((((-(((.........)))))).....(((((((((((......))))))))))))))-------- ( -23.00) >DroVir_CAF1 125199 109 + 1 -------GAGUUUGUCUGGACUGUGGACUGACUGACAGCUGCACUCAUUGAAGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCAGCA---AC -------.((((.((((.......)))).))))....(((((...........((((-(((.........)))))))....(((((((((((......)))))))))))))))).---.. ( -30.80) >DroPse_CAF1 85379 98 + 1 -------GAGA--GAG---AGAG-CGGUUGACUGACAGCUGCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUUCC-------- -------....--..(---((.(-((((((.....))))))).))).......((((-(((.........)))))))....(((((((((((......)))))))))))...-------- ( -27.20) >DroGri_CAF1 94726 118 + 1 UCUGUUUGGCUUUGACGGGUUUUUGGACUGACUGACAGCUGCACUCAUUGAGGCAAUUUUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGGAUCU--CAC ((.(((((((((..(.((((...(((.(((.....)))))).)))).)..))))(((((.....))))).))))).))..((((((((((((......)))))))))))).....--... ( -27.40) >DroWil_CAF1 110846 99 + 1 -------GAGGAUAU----GUAA-UGGCUGACUGACAGCUGCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGUU-------- -------...(((.(----((((-((((((.....)))))((.(((...)))))...-........)))))).)))..((((((((((((((......))))))))))))))-------- ( -27.00) >DroMoj_CAF1 131989 95 + 1 ------------UG------------UCUGACUGACAGCUGCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCAGCAUGAAC ------------((------------((.....))))(((((.(((...))).((((-(((.........)))))))....(((((((((((......))))))))))))))))...... ( -27.80) >consensus _______GAGA_UGAC___ACUG_GGGCUGACUGACAGCUGCACUCAUUGAGGCAAU_UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCA________ ............................(((((...((((((.(((...)))))).....)))..)))))..........((((((((((((......)))))))))))).......... (-18.57 = -18.90 + 0.33)
| Location | 18,231,992 – 18,232,092 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18231992 100 - 22407834 --------GGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUAUUUCAAUUUGUAACUUAAGCAA-AUUGCCUCAAUGAGUGCAGCUGUCAGUCAACCU-CAGU---GGCAUUCUA------- --------(((((((((((((........)))))))))).....(((((((.......))))-)))))).....((((((.((((...........-))))---.))))))..------- ( -28.60) >DroVir_CAF1 125199 109 - 1 GU---UGCUGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUAUUUCAAUUUGUAACUUAAGCAA-AUUGCUUCAAUGAGUGCAGCUGUCAGUCAGUCCACAGUCCAGACAAACUC------- ((---((((((((((((((((........))))))........((((((((.......))))-)))).......)))))))))((((.(.(........).)..)))))))..------- ( -25.50) >DroPse_CAF1 85379 98 - 1 --------GGAAUUUGCAUUUCUAAGAUAAAAUGCAAAUAUUUCAAUUUGUAACUUAAGCAA-AUUGCCUCAAUGAGUGCAGCUGUCAGUCAACCG-CUCU---CUC--UCUC------- --------((.((((((((((........))))))))))....((((((((.......))))-)))))).....(((.(.(((.((......)).)-))).---)))--....------- ( -22.00) >DroGri_CAF1 94726 118 - 1 GUG--AGAUCCAUUUGCAUUUCUAAGAUAAAAUGCAAAUAUUUCAAUUUGUAACUUAAGCAAAAUUGCCUCAAUGAGUGCAGCUGUCAGUCAGUCCAAAAACCCGUCAAAGCCAAACAGA .((--((((..((((((((((........)))))))))))))))).(((((..(((..((.....((((((...))).)))((((.....))))..........))..)))....))))) ( -21.90) >DroWil_CAF1 110846 99 - 1 --------AACAUUUGCAUUUCUAAGAUAAAAUGCAAAUAUUUCAAUUUGUAACUUAAGCAA-AUUGCCUCAAUGAGUGCAGCUGUCAGUCAGCCA-UUAC----AUAUCCUC------- --------...((((((((((........))))))))))....((((((((.......))))-)))).....(((((((..((((.....))))))-)).)----))......------- ( -21.20) >DroMoj_CAF1 131989 95 - 1 GUUCAUGCUGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUAUUUCAAUUUGUAACUUAAGCAA-AUUGCCUCAAUGAGUGCAGCUGUCAGUCAGA------------CA------------ ......(((((((((((((((........))))))........((((((((.......))))-)))).......))))))))).(((.....))------------).------------ ( -25.90) >consensus ________UGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUAUUUCAAUUUGUAACUUAAGCAA_AUUGCCUCAAUGAGUGCAGCUGUCAGUCAGCCC_CAAU___GACA_ACUC_______ ...........((((((((((........))))))))))........(((..(((..(((.....((((((...))).))))))...))))))........................... (-14.69 = -14.33 + -0.36)

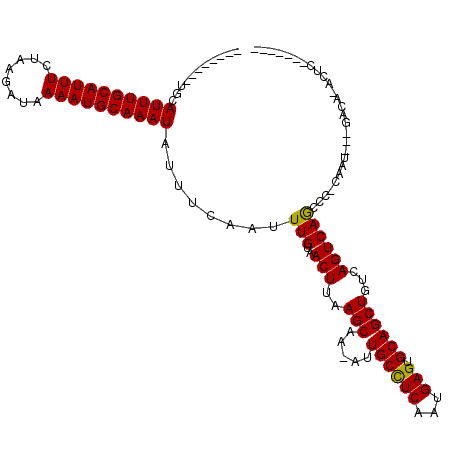
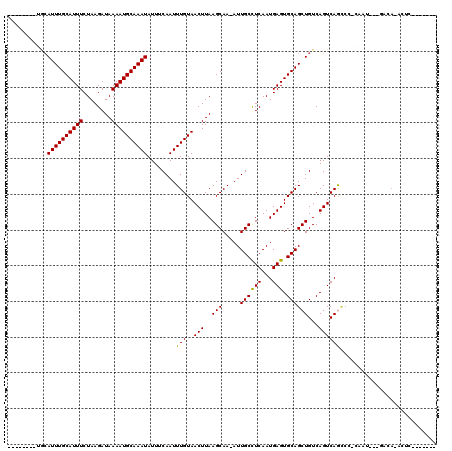
| Location | 18,232,021 – 18,232,116 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Mean single sequence MFE | -19.35 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18232021 95 + 22407834 GCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCC---------CUUUCUGCG------AGAUUUUCCGAAACU ...(((...((((((((-(((.........)))))))...((((((((((((......)))))))))))).)---------))).....)------))............. ( -20.70) >DroPse_CAF1 85406 93 + 1 GCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUUCC---------CCUGCACCC------CGCA-CCCCGGCA-C ((........(((((((-(((.........)))))))....(((((((((((......)))))))))))...---------)))((....------.)).-.....)).-. ( -18.90) >DroGri_CAF1 94766 109 + 1 GCACUCAUUGAGGCAAUUUUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGGAUCU--CACCCGCCCCUCCCCCACCCAACUUGCUGCUGCU ((........(((((....))))).(((..(((.(((....(((((((((((......)))))))))))((....--...))..............))).)))..))))). ( -21.20) >DroMoj_CAF1 132005 109 + 1 GCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCAGCAUGAACCCGCCCCCCACCCACCAACCCCGCC-CAGCU .......(((.((((((-(((.........))))))....((((((((((((......))))))))))))................................)))-))).. ( -18.80) >DroAna_CAF1 69498 84 + 1 GCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCU---------CUUUCUGCG------AGAU----------- (((......(((.((((-(((.........)))))))...((((((((((((......))))))))))))))---------)....))).------....----------- ( -20.20) >DroPer_CAF1 85986 85 + 1 GCACUCAUUGAGGCAAU-UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUUCC---------CCCGCACCC------CGCA----------C ((.(((...))).((((-(((.........)))))))....(((((((((((......)))))))))))...---------...))....------....----------. ( -16.30) >consensus GCACUCAUUGAGGCAAU_UUGCUUAAGUUACAAAUUGAAAUAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCC_________CCCCCACCC______AGAA___CC_____U ..........(((((....)))))................((((((((((((......))))))))))))......................................... (-13.72 = -14.05 + 0.33)

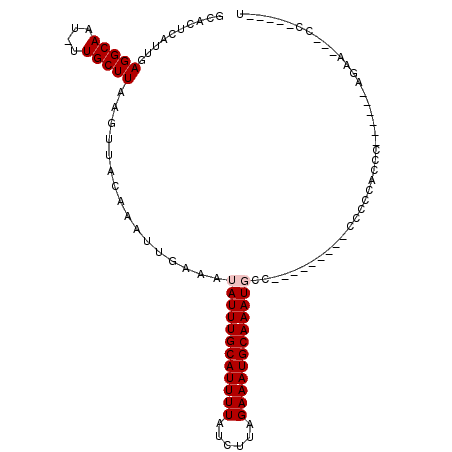
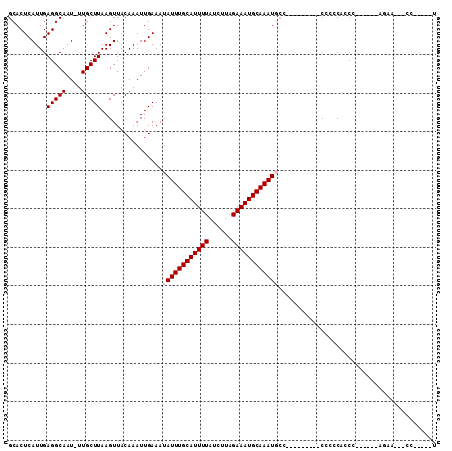
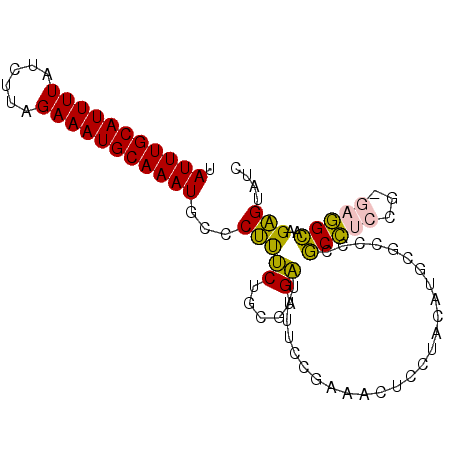
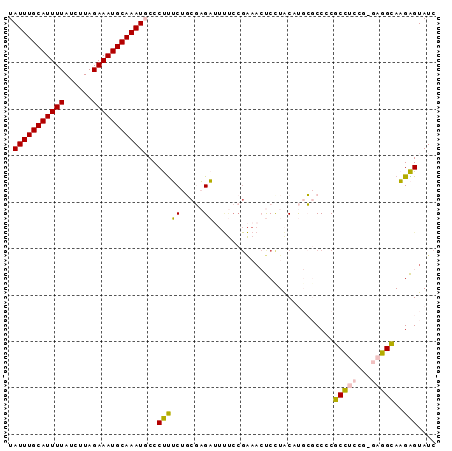
| Location | 18,232,060 – 18,232,151 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -16.05 |
| Energy contribution | -14.80 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18232060 91 + 22407834 UAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCCCUUUCUGCGAGAUUUUCCGAAACUCCUACAUGCGCCCCGCCUCCU-GAGGCAAGAGUAUC ((((((((((((......))))))))))))....((((..(((.(((....)))))).............((((...-.)))).)))).... ( -19.20) >DroPse_CAF1 85445 90 + 1 UAUUUGCAUUUUAUCUUAGAAAUGCAAAUUCCCCUGCACCCCGCA-CCCCGGCA-CCCUGCACCGCCCCCACGUACGGGCUGUACGGGUAUC .(((((((((((......))))))))))).(((.((((.((((.(-(...(((.-.........))).....)).)))).)))).))).... ( -28.80) >DroSec_CAF1 75217 91 + 1 UAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCCCUUUCUGCGAGAUUUUCCGAAACUCUUACAUGCGCCCCGCCUCCU-GAGGCAAGAGUAUC ((((((((((((......))))))))))))....................((.((((((.(....)....((((...-.)))))))))).)) ( -20.60) >DroEre_CAF1 75258 91 + 1 UAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCCCUUUCUGCGAGAUUUUCCGAAACUCCUACAUGCGCCCCGCCUCCG-GAGGCAAGAGUAUC ((((((((((((......))))))))))))....((((..(((.(((....))))))........((((((....))-).))).)))).... ( -21.80) >DroYak_CAF1 79645 91 + 1 UAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCCCUUUCUGCGAGAUUUUCCGAAACUCCUACAUGCGCCCCGCCUCCU-GAGGCAAGAGUAUC ((((((((((((......))))))))))))....((((..(((.(((....)))))).............((((...-.)))).)))).... ( -19.20) >DroPer_CAF1 86025 82 + 1 UAUUUGCAUUUUAUCUUAGAAAUGCAAAUUCCCCCGCACCCCGCA----------CCCUACACCGCCCCCACGUACGGGCUGUACGGGUAUC .(((((((((((......)))))))))))......((.....))(----------(((((((..((((........)))))))).))))... ( -26.10) >consensus UAUUUGCAUUUUAUCUUAGAAAUGCAAAUGCCCUUUCUGCGAGAUUUUCCGAAACUCCUACAUGCGCCCCGCCUCCG_GAGGCAAGAGUAUC .(((((((((((......)))))))))))...(((((.....))..........................(((((...)))))..))).... (-16.05 = -14.80 + -1.25)

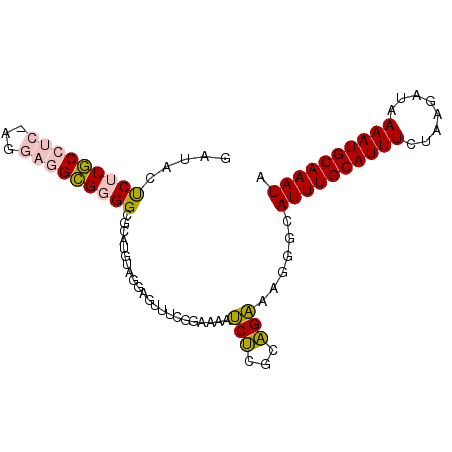
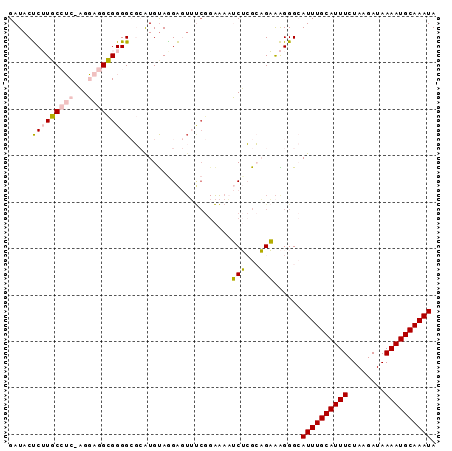
| Location | 18,232,060 – 18,232,151 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -16.12 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18232060 91 - 22407834 GAUACUCUUGCCUC-AGGAGGCGGGGCGCAUGUAGGAGUUUCGGAAAAUCUCGCAGAAAGGGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUA ......(((((((.-...)))))))((.(.((..(((...........)))..))....).))((((((((((........)))))))))). ( -24.80) >DroPse_CAF1 85445 90 - 1 GAUACCCGUACAGCCCGUACGUGGGGGCGGUGCAGGG-UGCCGGGG-UGCGGGGUGCAGGGGAAUUUGCAUUUCUAAGAUAAAAUGCAAAUA ....(((.(.((.(((((((.(.((.((........)-).)).).)-)))))).)).).))).((((((((((........)))))))))). ( -39.20) >DroSec_CAF1 75217 91 - 1 GAUACUCUUGCCUC-AGGAGGCGGGGCGCAUGUAAGAGUUUCGGAAAAUCUCGCAGAAAGGGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUA ......(((((((.-...)))))))((.(.((..(((...........)))..))....).))((((((((((........)))))))))). ( -24.80) >DroEre_CAF1 75258 91 - 1 GAUACUCUUGCCUC-CGGAGGCGGGGCGCAUGUAGGAGUUUCGGAAAAUCUCGCAGAAAGGGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUA ((.(((((((((((-((....))))).)))....))))).))..........((.......))((((((((((........)))))))))). ( -26.30) >DroYak_CAF1 79645 91 - 1 GAUACUCUUGCCUC-AGGAGGCGGGGCGCAUGUAGGAGUUUCGGAAAAUCUCGCAGAAAGGGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUA ......(((((((.-...)))))))((.(.((..(((...........)))..))....).))((((((((((........)))))))))). ( -24.80) >DroPer_CAF1 86025 82 - 1 GAUACCCGUACAGCCCGUACGUGGGGGCGGUGUAGGG----------UGCGGGGUGCGGGGGAAUUUGCAUUUCUAAGAUAAAAUGCAAAUA ....(((((((..(((((((.(....((...)).).)----------)))))))))))))...((((((((((........)))))))))). ( -32.20) >consensus GAUACUCUUGCCUC_AGGAGGCGGGGCGCAUGUAGGAGUUUCGGAAAAUCUCGCAGAAAGGGCAUUUGCAUUUCUAAGAUAAAAUGCAAAUA .....(((((((((...)))))))))......................(((...)))......((((((((((........)))))))))). (-16.12 = -15.90 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:25 2006