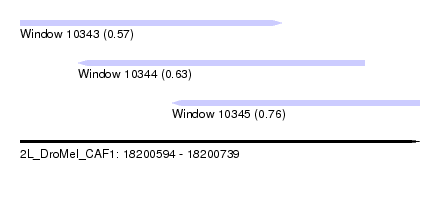
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,200,594 – 18,200,739 |
| Length | 145 |
| Max. P | 0.760213 |

| Location | 18,200,594 – 18,200,689 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -22.01 |
| Energy contribution | -23.17 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18200594 95 + 22407834 ------GCU-CGCAUUGCUGUUGCUG---------CUGCUGUGAUUGCUGGUCGUGGUGCACUUGAGCAAUGUGGCAGUGCACUCAUCCAAUGCAGACUUAGUGCACUCCU ------((.-(((((((((...(.((---------(.((..((((.....))))..))))))...)))))))))))((((((((..((.......))...))))))))... ( -35.40) >DroVir_CAF1 66465 102 + 1 GAUCCGGCUUGGCAUUGCUGUUGCUGCCGCCGCCGCC---------GCCGCUGUUUGCGCACUUCAGCAAUGUGGCCGCGCAUUCGUCCAGAGCCGUUUUGGUGCAAUCCU ((..((((((((((..((....))))))...((.((.---------(((((...((((........)))).))))).)))).........))))))..))((......)). ( -36.30) >DroSec_CAF1 43562 95 + 1 ------GCU-CGCAUUGCUGUUGCUG---------CUGCUGUGAUUGCUGGUCGUGGUGCACUUGAGCAAUGUGGCAGUGCACUCAUCCAAUGCAGACUUAGUGCACUCCU ------((.-(((((((((...(.((---------(.((..((((.....))))..))))))...)))))))))))((((((((..((.......))...))))))))... ( -35.40) >DroSim_CAF1 44171 95 + 1 ------GCU-CGCAUUGCUGUUGCUG---------CUGCUGUGAUUGCUGGUCGUGGUGCACUUGAGCAAUGUGGCAGUGCACUCAUCCAACGCAGACUUAGUGCACUCCU ------((.-(((((((((...(.((---------(.((..((((.....))))..))))))...)))))))))))((((((((..((.......))...))))))))... ( -35.40) >DroEre_CAF1 43745 95 + 1 ------GCU-UGCAUUGCUGUUGCUG---------CUGCUGUGAUUGCUGGUCGUGGUGCACUUGAGCAAUGUGGCAGUGCACUCGUCCAAUGCAGACUUGGUGCACUCUU ------(((-.((((((((...(.((---------(.((..((((.....))))..))))))...)))))))))))((((((((.(((.......)))..))))))))... ( -35.40) >consensus ______GCU_CGCAUUGCUGUUGCUG_________CUGCUGUGAUUGCUGGUCGUGGUGCACUUGAGCAAUGUGGCAGUGCACUCAUCCAAUGCAGACUUAGUGCACUCCU ......(((..((((((((..(((.............((..((((.....))))..)))))....)))))))))))((((((((.(..(......)..).))))))))... (-22.01 = -23.17 + 1.16)



| Location | 18,200,615 – 18,200,719 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -25.17 |
| Energy contribution | -25.48 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18200615 104 - 22407834 UGGCGCAGCUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACUAAGUCUGCAUUGGAUGAGUGCACUGCCACAUUGCUCAAGUGCACCACGACCAGC------AAUCACAGC .......((((((((((((....))))))).((((((((((..((((.....)))).)))))))).))..((((((...(((...)))....)))------))).))))) ( -38.50) >DroPse_CAF1 47203 110 - 1 UGGCUCAAUUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACCAAAUCCGCACUGGACGAGUGCACGGCCACACUGCUCCAGUGCACCACCACCAGCACCGCCAAUCAGAGC ((((....((.((((((((....)))))))).))((((.........((((((((..((((........))))..))))))))..........)))).))))........ ( -40.21) >DroSim_CAF1 44192 104 - 1 UGGCGCAGCUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACUAAGUCUGCGUUGGAUGAGUGCACUGCCACAUUGCUCAAGUGCACCACGACCAGC------AAUCACAGC .......((((((((((((....))))))).((((((((((..((((.....)))).)))))))).))..((((((...(((...)))....)))------))).))))) ( -38.50) >DroEre_CAF1 43766 104 - 1 UGUCGCAGCUGUGCAGCAGCGAUCUGUUGCAAGAGUGCACCAAGUCUGCAUUGGACGAGUGCACUGCCACAUUGCUCAAGUGCACCACGACCAGC------AAUCACAGC .......((((((((((((....)))))))...(((((((...((((.....))))..))))))).....((((((...(((...)))....)))------))).))))) ( -36.00) >DroWil_CAF1 66886 101 - 1 UGGCUCAAUUGUGUAGCAGUGAUCUGUUACAAGAUUGCACUAAAUCAGCUUUAGAUGAAUGCACUGCCACGCUAUUGCAAUGCACCA---CCAGC------AAUGGCAGC ((((.(((((.((((((((....)))))))).)))))(((((((.....))))).))........)))).((((((((.........---...))------))))))... ( -30.90) >DroPer_CAF1 47032 110 - 1 UGGCUCAAUUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACCAAAUCCGCACUGAACGAGUGCACGGUCACACUGCUCCAGUGCACCACCACCAGCACCGCCAAUCAGAGC ((((....((.((((((((....)))))))).))((((.........(((((.....)))))..(((..(((((...))))).))).......)))).))))........ ( -38.30) >consensus UGGCGCAACUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACCAAAUCUGCAUUGGACGAGUGCACUGCCACACUGCUCAAGUGCACCACGACCAGC______AAUCACAGC (((((......((((((((....))))))))...((((((...((((.....))))..)))))))))))...(((......))).......................... (-25.17 = -25.48 + 0.31)

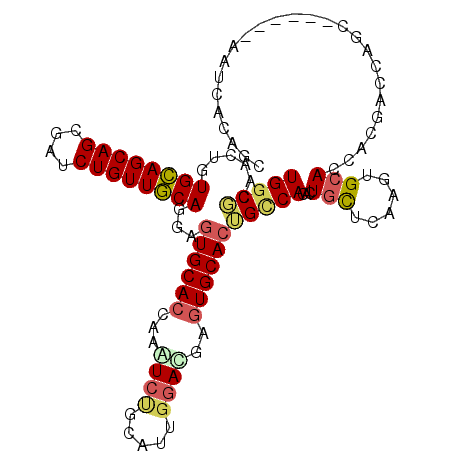
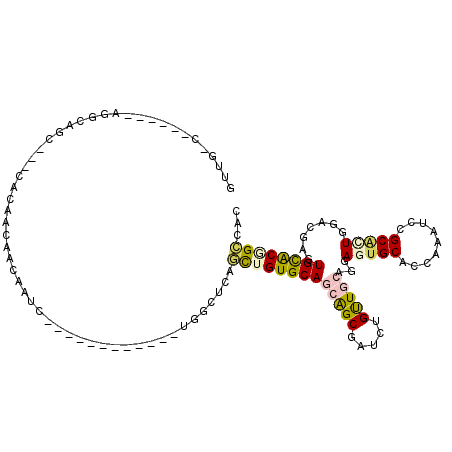
| Location | 18,200,649 – 18,200,739 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.49 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -16.54 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18200649 90 - 22407834 ------------AAGCAGC---CACAAUAACAACU------------UGGCGCAGCUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACUAAGUCUGCAUUGGAUGAGUGCACUGCCAC ------------.(((.((---((...........------------))))...))).((((((((....))))))))((((((((((..((((.....)))).)))))))).)).. ( -34.90) >DroVir_CAF1 66527 105 - 1 AUUGCC------------CAAGAGCAACAACAAUCUGGCCCUGGGUAUGGCACAGCUGUGCAACGGCGAUCUGCUUGAGGAUUGCACCAAAACGGCUCUGGACGAAUGCGCGGCCAC .((((.------------.....))))........(((((.((.((.((.((.((((((......(((((((......)))))))......)))))).))..)).)).)).))))). ( -33.60) >DroPse_CAF1 47243 102 - 1 GUUGACCUUCUCGGGCAGC---CACAACAACAAUC------------UGGCUCAAUUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACCAAAUCCGCACUGGACGAGUGCACGGCCAC ((((.(((....)))))))---.............------------(((((...((.((((((((....)))))))).))((((((....(((.....)))...))))))))))). ( -35.60) >DroSim_CAF1 44226 90 - 1 ------------AAGCAGC---CACAACAACAACU------------UGGCGCAGCUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACUAAGUCUGCGUUGGAUGAGUGCACUGCCAC ------------.(((.((---((...........------------))))...))).((((((((....))))))))((((((((((..((((.....)))).)))))))).)).. ( -34.90) >DroMoj_CAF1 68610 117 - 1 GUUGCCAUUAGCUGUCAACAAGAGCAACAACAAUUUGGCAAUGGGCAUGGCUCAGCUGUGCAGCAGCGAUCUGCUAGAGGACUGCACCAAGACGGCGCUCGAUGAUUGUGCAGCGAC (((((.....((((((.................(((((((.(((((...)))))((((.....))))....)))))))((......))..))))))((.(((...))).)).))))) ( -35.50) >DroPer_CAF1 47072 102 - 1 GUUGACCUUCUCGGGCAGC---CACAACAACAAUC------------UGGCUCAAUUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACCAAAUCCGCACUGAACGAGUGCACGGUCAC ..(((((.....(((((((---((...........------------))))....((.((((((((....)))))))).)).))).))......(((((.....)))))..))))). ( -36.10) >consensus GUUG_C______AGGCAGC___CACAACAACAAUC____________UGGCUCAGCUGUGCAGCAGCGAUCUGUUGCAGGAGUGCACCAAAUCCGCACUGGACGAGUGCACGGCCAC ......................................................(((((((((((((.....)))))...(((((.........))))).......))))))))... (-16.54 = -16.68 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:14 2006