| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,200,177 – 18,200,297 |
| Length | 120 |
| Max. P | 0.743948 |

| Location | 18,200,177 – 18,200,297 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.82 |
| Mean single sequence MFE | -46.70 |
| Consensus MFE | -36.04 |
| Energy contribution | -36.85 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
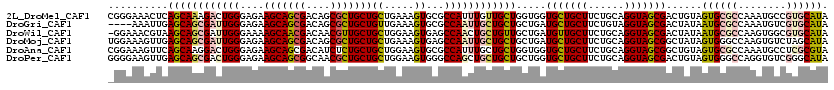
>2L_DroMel_CAF1 18200177 120 + 22407834 CGGGAAACUCAGCAAAGACUGGGAGAAGCAGCGACAGCGCUGCUGCUGAAAGUGCGCCAUUUGUUGCUGGUGGUGCUGCUUCUGCAGGUAGCGACUGUAGUGCGCCAAAUGCCGUGCAUA (((....(((((......)))))...(((((((....))))))).)))...(..((.((((((.(((..(..((((((((......)))))).))..)...))).)))))).))..)... ( -46.60) >DroGri_CAF1 49047 116 + 1 ----AAAUUGAGCAGCGAUUGGGAGAAGCAGCGACAGCGCUGCUGUUGAAAGUGCGCCAAUUGCUGCUGCUGAUGCUGCUUCUGUAGGUAGCGACUAUAAUGCGCCAAAUGUCGUGCAUA ----......((((((((((((....(((((((....)))))))((.......)).)))))))))))).....(((((((......)))))))......((((((........)))))). ( -46.10) >DroWil_CAF1 66506 119 + 1 -GGAAACGUAAGCAGCGAUUGGGAAAAGCAACGACAACGUUGCUGCUGGAAGUGAGCCAACUGCUGUUGCUGAUGUUGCUUCUGCAGGUAGCGACUAUAAUGCGCCAAGUGGCGUGCAUA -(....)....((((((.((((....(((((((....)))))))((.....))...)))).))))(((((((...((((....)))).)))))))......(((((....)))))))... ( -40.80) >DroMoj_CAF1 68183 120 + 1 UGGAAAGUUGAGCAGCGAUUGGGAGAAGCAGCGACAGCGCUGCUGCUGAAAGUGAGCCAAUUGCUGCUGCUGAUGCUGCUUCUGCAGGUAGCGGCUAUAGUGGGCCAAGUGUCUAGCAUA ..........((((((((((((....(((((((....)))))))((.....))...))))))))))))((((((((((((......))))))((((......))))....))).)))... ( -50.50) >DroAna_CAF1 38973 120 + 1 CGGAAAGUUCAGCAAGGACUGGGAGAAGCAGCGACAUCUCUGCUGCUGGAAGUGCGCCAUUUGCUGCUGGUGGUGCUGCUUCUGCAGGUAGCGGCUGUAGUGCGCCAAAUGCCUCGCGUA .....(((((.....)))))......(((((((.......)))))))((((((((((((((.......)))))))).))))))(((((((..(((........)))...))))).))... ( -43.50) >DroPer_CAF1 46591 120 + 1 GGGGAAGUUGAGCAGCGACUGGGAGAAGCAGCGGCAACGCUGCUGCUGGAAGUGGGCCAGCUGCUGCUGCUGGUGCUGCUUCUGCAGGUAGCGACUGUAGUGGGCCAGGUGUCGGGCAUA .........(((((((.((..(.((.(((((((....)))))))(((((.......))))).....)).)..)))))))))((((((.......))))))...(((........)))... ( -52.70) >consensus _GGAAAGUUGAGCAGCGACUGGGAGAAGCAGCGACAGCGCUGCUGCUGAAAGUGCGCCAAUUGCUGCUGCUGAUGCUGCUUCUGCAGGUAGCGACUAUAGUGCGCCAAAUGUCGUGCAUA ..........((((((((((((....(((((((....)))))))((.....))...)))))))))))).....(((((((......)))))))......((((((........)))))). (-36.04 = -36.85 + 0.81)

| Location | 18,200,177 – 18,200,297 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.82 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -26.67 |
| Energy contribution | -26.28 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18200177 120 - 22407834 UAUGCACGGCAUUUGGCGCACUACAGUCGCUACCUGCAGAAGCAGCACCACCAGCAACAAAUGGCGCACUUUCAGCAGCAGCGCUGUCGCUGCUUCUCCCAGUCUUUGCUGAGUUUCCCG ...((.(((((..(((((.((....))))))).(((.(((((((((..(.(((........))).)......((((......))))..)))))))))..)))....))))).))...... ( -36.70) >DroGri_CAF1 49047 116 - 1 UAUGCACGACAUUUGGCGCAUUAUAGUCGCUACCUACAGAAGCAGCAUCAGCAGCAGCAAUUGGCGCACUUUCAACAGCAGCGCUGUCGCUGCUUCUCCCAAUCGCUGCUCAAUUU---- ...((((((....((((((......).))))).....(((((((((..((((.((.((..((((.......))))..)).))))))..))))))))).....))).))).......---- ( -37.70) >DroWil_CAF1 66506 119 - 1 UAUGCACGCCACUUGGCGCAUUAUAGUCGCUACCUGCAGAAGCAACAUCAGCAACAGCAGUUGGCUCACUUCCAGCAGCAACGUUGUCGUUGCUUUUCCCAAUCGCUGCUUACGUUUCC- ...(((.(((....)))((.........((.....))(((((((((..((((....((.(((((.......))))).))...))))..))))))))).......)))))..........- ( -32.50) >DroMoj_CAF1 68183 120 - 1 UAUGCUAGACACUUGGCCCACUAUAGCCGCUACCUGCAGAAGCAGCAUCAGCAGCAGCAAUUGGCUCACUUUCAGCAGCAGCGCUGUCGCUGCUUCUCCCAAUCGCUGCUCAACUUUCCA ..((((.((.....(((........))).....((((....))))..))))))(((((.(((((((.......)))(((((((....))))))).....)))).)))))........... ( -36.70) >DroAna_CAF1 38973 120 - 1 UACGCGAGGCAUUUGGCGCACUACAGCCGCUACCUGCAGAAGCAGCACCACCAGCAGCAAAUGGCGCACUUCCAGCAGCAGAGAUGUCGCUGCUUCUCCCAGUCCUUGCUGAACUUUCCG ...(((((((....(((........)))((.....))(((((((((..((((.((.((.....))((.......)).)).).).))..)))))))))....).))))))........... ( -35.90) >DroPer_CAF1 46591 120 - 1 UAUGCCCGACACCUGGCCCACUACAGUCGCUACCUGCAGAAGCAGCACCAGCAGCAGCAGCUGGCCCACUUCCAGCAGCAGCGUUGCCGCUGCUUCUCCCAGUCGCUGCUCAACUUCCCC ...((.((((..(((........)))..((.....))(((((((((..((((.((.((.(((((.......))))).)).))))))..)))))))))....))))..))........... ( -40.80) >consensus UAUGCACGACACUUGGCGCACUACAGUCGCUACCUGCAGAAGCAGCACCAGCAGCAGCAAUUGGCGCACUUCCAGCAGCAGCGCUGUCGCUGCUUCUCCCAAUCGCUGCUCAACUUUCC_ ..(((.........(((........)))((.....))....)))......(.((((((.(((((.........((.(((((((....))))))).)).))))).)))))))......... (-26.67 = -26.28 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:12 2006