| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,189,968 – 18,190,072 |
| Length | 104 |
| Max. P | 0.859911 |

| Location | 18,189,968 – 18,190,072 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
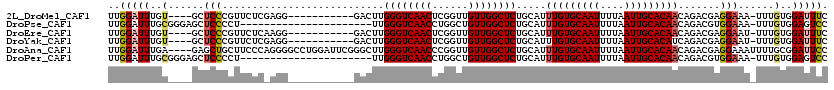
>2L_DroMel_CAF1 18189968 104 + 22407834 UUGGAUUUGU----GCUCCCGUUCUCGAGG-----------GACUUGGGUCAACUCGGUUGUUGGCUCUGCAUUUGUGCAAUUUUAAUUGCACAACAGACGAGGAAA-UUUGUGGAUUUC .....(((((----((((((........))-----------))...((((((((......)))))))).))..(((((((((....)))))))))...)))))((((-((....)))))) ( -31.40) >DroPse_CAF1 34835 97 + 1 UUGGAUUUGCGGGAGCUCCCCU----------------------UUGGGUCAACCUGGCUGUUGGCUCUGCAUUUGUGCAAUUUUAAUUGCACAACAGACGUGGAAA-UUUGUGGAGUCC ..(((((((((((....)))..----------------------..((((((((......)))))))).))..(((((((((....)))))))))((.(.((....)-).).)))))))) ( -31.40) >DroEre_CAF1 33029 104 + 1 UUGGAUUUGU----GCUCCCGUUCUCAAGG-----------GACUUGGGUCAACUCGGUUGUUGGCUCUGCAUUUGUGCAAUUUUAAUUGCACAACAGACGAGGAAU-UUUGUGGAUUUC ..(((((..(----..((((........))-----------))((((.((((((......))))))((((...(((((((((....)))))))))))))))))....-...)..))))). ( -31.30) >DroYak_CAF1 32789 104 + 1 UUGGAUUUGU----GCUCCCGUUCUCGAGG-----------GACUUGGGUCAACUCGGUUGUUGGCUCUGCAUUUGUGCAAUUUUAAUUGCACAUCAGACGAGGAAU-UUUGUGGAUUUC ..(((((..(----..((((........))-----------))((((.((((((......))))))((((....((((((((....)))))))).))))))))....-...)..))))). ( -30.80) >DroAna_CAF1 29532 116 + 1 UUGGAUUUGA----GAGCUGCUUCCCAGGGGCCUGGAUUCGGGCUUGGGUCAACCCGGUUGUUGGCUCUGCAUUUGUGCAAUUUUAAUUGCACAACAGACGAGGAAAUUUUGCGGAUUCC ..((((((((----(((((..(..((..(((((((....)))))))(((....)))))..)..))))))....(((((((((....))))))))).................))))))). ( -39.40) >DroPer_CAF1 34715 97 + 1 UUGGAUUUGCGGGAGCUCCCCU----------------------UUGGGUCAACCUGGCUGUUGGCUCUGCAUUUGUGCAAUUUUAAUUGCACAACAGACGUGGAAA-UUUGUGGAGUCC ..(((((((((((....)))..----------------------..((((((((......)))))))).))..(((((((((....)))))))))((.(.((....)-).).)))))))) ( -31.40) >consensus UUGGAUUUGU____GCUCCCCUUCUC_AGG___________GACUUGGGUCAACCCGGUUGUUGGCUCUGCAUUUGUGCAAUUUUAAUUGCACAACAGACGAGGAAA_UUUGUGGAUUCC ..(((((..(......(((...........................((((((((......)))))))).....(((((((((....))))))))).......)))......)..))))). (-23.10 = -23.38 + 0.28)

| Location | 18,189,968 – 18,190,072 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -17.65 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
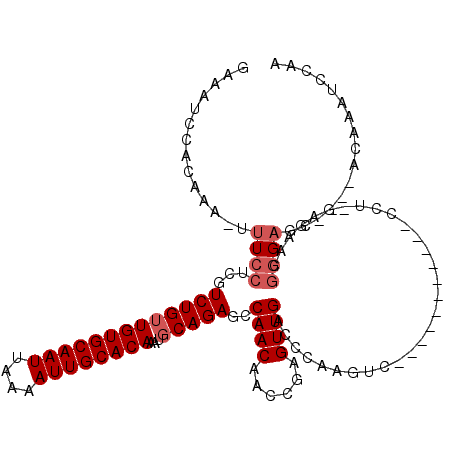
>2L_DroMel_CAF1 18189968 104 - 22407834 GAAAUCCACAAA-UUUCCUCGUCUGUUGUGCAAUUAAAAUUGCACAAAUGCAGAGCCAACAACCGAGUUGACCCAAGUC-----------CCUCGAGAACGGGAGC----ACAAAUCCAA (((((......)-))))(((.(((((((((((((....))))))))...)))))..........(((..(((....)))-----------.))))))....(((..----.....))).. ( -25.10) >DroPse_CAF1 34835 97 - 1 GGACUCCACAAA-UUUCCACGUCUGUUGUGCAAUUAAAAUUGCACAAAUGCAGAGCCAACAGCCAGGUUGACCCAA----------------------AGGGGAGCUCCCGCAAAUCCAA (((.........-............(((((((((....))))))))).(((.((((((((......)))).(((..----------------------..))).))))..)))..))).. ( -28.40) >DroEre_CAF1 33029 104 - 1 GAAAUCCACAAA-AUUCCUCGUCUGUUGUGCAAUUAAAAUUGCACAAAUGCAGAGCCAACAACCGAGUUGACCCAAGUC-----------CCUUGAGAACGGGAGC----ACAAAUCCAA ............-........(((((((((((((....))))))))...)))))((((((......)))).......((-----------((........))))))----.......... ( -23.70) >DroYak_CAF1 32789 104 - 1 GAAAUCCACAAA-AUUCCUCGUCUGAUGUGCAAUUAAAAUUGCACAAAUGCAGAGCCAACAACCGAGUUGACCCAAGUC-----------CCUCGAGAACGGGAGC----ACAAAUCCAA ............-.((((.((((((.((((((((....))))))))....)))).........((((..(((....)))-----------.))))....)))))).----.......... ( -23.90) >DroAna_CAF1 29532 116 - 1 GGAAUCCGCAAAAUUUCCUCGUCUGUUGUGCAAUUAAAAUUGCACAAAUGCAGAGCCAACAACCGGGUUGACCCAAGCCCGAAUCCAGGCCCCUGGGAAGCAGCUC----UCAAAUCCAA (((......................(((((((((....))))))))).((.(((((.......((((((......))))))..(((((....))))).....))))----)))..))).. ( -37.50) >DroPer_CAF1 34715 97 - 1 GGACUCCACAAA-UUUCCACGUCUGUUGUGCAAUUAAAAUUGCACAAAUGCAGAGCCAACAGCCAGGUUGACCCAA----------------------AGGGGAGCUCCCGCAAAUCCAA (((.........-............(((((((((....))))))))).(((.((((((((......)))).(((..----------------------..))).))))..)))..))).. ( -28.40) >consensus GAAAUCCACAAA_UUUCCUCGUCUGUUGUGCAAUUAAAAUUGCACAAAUGCAGAGCCAACAACCGAGUUGACCCAAGUC___________CCU_GAGAACGGGAGC____ACAAAUCCAA ..............((((...(((((((((((((....))))))))...)))))..((((......))))..............................))))................ (-17.65 = -18.32 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:02 2006