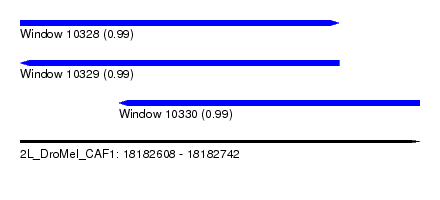
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,182,608 – 18,182,742 |
| Length | 134 |
| Max. P | 0.994739 |

| Location | 18,182,608 – 18,182,715 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.84 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18182608 107 + 22407834 AGGGGGCACAAGGUGCCGGCAGAUCCCUUCUGCCCACCAGAACCCCAAAGUCACCCAAAACCCCCGCAGAACGGGCACUUAAGUACACAACUACCCUCUCCAGAGUU .((((......((((..((((((.....))))))))))....))))................((((.....))))....................(((....))).. ( -29.70) >DroSec_CAF1 25365 107 + 1 AGAGGGCACAAGGUGCCGGCAGAUCCCUUCUGCCCACCAGAACCCCAAAGUCACCCAAAACCCCCGCAGAACGGGCACUUAAGUACACAACCACCCUCUGCAGAGUU ((((((.....((((..((((((.....))))))))))........................((((.....))))..................))))))........ ( -30.40) >DroSim_CAF1 26005 107 + 1 AGAGGGCACAAGGUGCCGGCAGAUCCCUUCUGCCCACCAGAACCCCAAAGUCACCCAAAACCCCCGCAGAACGGGCACUUAAGUACACAACCACCCUCUGCAGAGUU ((((((.....((((..((((((.....))))))))))........................((((.....))))..................))))))........ ( -30.40) >DroEre_CAF1 25465 106 + 1 AGAGGGCACAAGGUGCCAGCAGU-CCCUUCUGCCCGCCAGAGCCCCAAAGUCGCCCAAAACUCCCGCAGAACGGGCACUUAAGUACUCAACUACCCUCUCCAGAAUU ((((((.....((((...((((.-.....)))).)))).(((..(..((((.((((................))))))))..)..))).....))))))........ ( -31.69) >DroYak_CAF1 25398 107 + 1 AGAGGGCACAAGGUGCCAGCAGUUCCCUUCUGCCCGCCAGAGCCCCAAAGUCACCCAAAACUGCCGCAGAACGGGCACUUAAGUACACAACUACCCUCUCCAGAGCU ((((((......((((....(((.(((((((((..((....)).....(((........)))...)))))).))).)))...)))).......))))))........ ( -27.72) >consensus AGAGGGCACAAGGUGCCGGCAGAUCCCUUCUGCCCACCAGAACCCCAAAGUCACCCAAAACCCCCGCAGAACGGGCACUUAAGUACACAACUACCCUCUCCAGAGUU ((((((.....((((..((((((.....))))))))))........................((((.....))))..................))))))........ (-25.24 = -25.84 + 0.60)

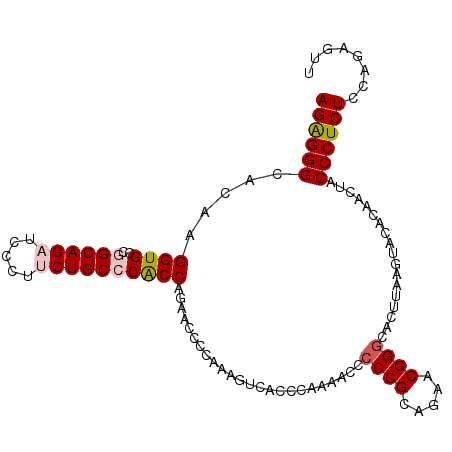
| Location | 18,182,608 – 18,182,715 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -44.48 |
| Consensus MFE | -37.90 |
| Energy contribution | -36.94 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18182608 107 - 22407834 AACUCUGGAGAGGGUAGUUGUGUACUUAAGUGCCCGUUCUGCGGGGGUUUUGGGUGACUUUGGGGUUCUGGUGGGCAGAAGGGAUCUGCCGGCACCUUGUGCCCCCU ..(((((.((((((((.(((......))).)))))..))).))))).....((....))..(((((...((((((((((.....))))))..))))....))))).. ( -44.90) >DroSec_CAF1 25365 107 - 1 AACUCUGCAGAGGGUGGUUGUGUACUUAAGUGCCCGUUCUGCGGGGGUUUUGGGUGACUUUGGGGUUCUGGUGGGCAGAAGGGAUCUGCCGGCACCUUGUGCCCUCU ..((((((((((((((.(((......))).)))))..))))))))).....((....))..(((((...((((((((((.....))))))..))))....))))).. ( -45.90) >DroSim_CAF1 26005 107 - 1 AACUCUGCAGAGGGUGGUUGUGUACUUAAGUGCCCGUUCUGCGGGGGUUUUGGGUGACUUUGGGGUUCUGGUGGGCAGAAGGGAUCUGCCGGCACCUUGUGCCCUCU ..((((((((((((((.(((......))).)))))..))))))))).....((....))..(((((...((((((((((.....))))))..))))....))))).. ( -45.90) >DroEre_CAF1 25465 106 - 1 AAUUCUGGAGAGGGUAGUUGAGUACUUAAGUGCCCGUUCUGCGGGAGUUUUGGGCGACUUUGGGGCUCUGGCGGGCAGAAGGG-ACUGCUGGCACCUUGUGCCCUCU ......((((.(((((.((((....)))).))))).))))((.(((((((..((....))..))))))).))(((((.((((.-.(.....)..)))).)))))... ( -45.20) >DroYak_CAF1 25398 107 - 1 AGCUCUGGAGAGGGUAGUUGUGUACUUAAGUGCCCGUUCUGCGGCAGUUUUGGGUGACUUUGGGGCUCUGGCGGGCAGAAGGGAACUGCUGGCACCUUGUGCCCUCU (((((..(((.(((((.(((......))).)))))(..(.((....))...)..)..)))..)))))..((.(((((.((((...(....)...)))).))))).)) ( -40.50) >consensus AACUCUGGAGAGGGUAGUUGUGUACUUAAGUGCCCGUUCUGCGGGGGUUUUGGGUGACUUUGGGGUUCUGGUGGGCAGAAGGGAUCUGCCGGCACCUUGUGCCCUCU ..(((((.((((((((.(((......))).)))))..))).))))).....((....))..(((((.(..((.(((((.......))))).)).....).))))).. (-37.90 = -36.94 + -0.96)

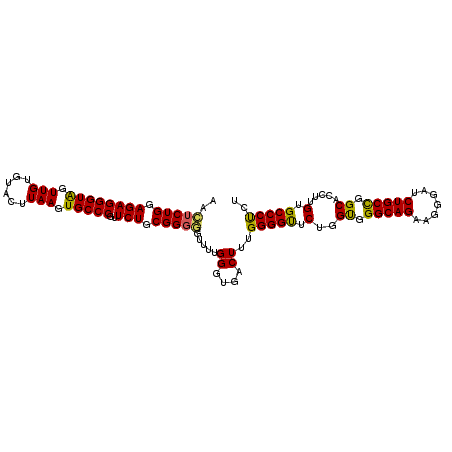

| Location | 18,182,641 – 18,182,742 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -38.71 |
| Consensus MFE | -23.91 |
| Energy contribution | -24.37 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18182641 101 - 22407834 CCCAGUGUUUGAAGGGAAUCCGUCC--AGAACUCUGGAGAGGGUAGUUGUGUACUUAAGUGCCCGUUCUGCGGGGG-UUUUGGGUGACUUUGGGGUUCUGGUGG .((((..(((.((((.....(..((--((((((((.(((((((((.(((......))).)))))..))).).))))-))))))..).)))).)))..))))... ( -37.40) >DroPse_CAF1 27639 81 - 1 ------------GGGGAAA-UAUGGUGGGAGGGCAAGGCAAGGCAGUUAUGUACUUAAGUGCCCUUGCUGUGGGCGGCACCGUGUG-CUUUGUGC--------- ------------((((..(-((((((..(((((((.......(((....))).......)))))))((((....))))))))))).-))))....--------- ( -26.74) >DroSec_CAF1 25398 101 - 1 ACCAGUGCUUGAAGGGAAUCCGUCC--AGAACUCUGCAGAGGGUGGUUGUGUACUUAAGUGCCCGUUCUGCGGGGG-UUUUGGGUGACUUUGGGGUUCUGGUGG (((((.((((.((((.....(..((--((((((((((((((((((.(((......))).)))))..))))).))))-))))))..).)))).)))).))))).. ( -44.60) >DroSim_CAF1 26038 101 - 1 CCCAGUGCUUGAAGGGAAUCCGUCC--AAAACUCUGCAGAGGGUGGUUGUGUACUUAAGUGCCCGUUCUGCGGGGG-UUUUGGGUGACUUUGGGGUUCUGGUGG .((((.((((.((((.....(..((--((((((((((((((((((.(((......))).)))))..))))).))))-))))))..).)))).)))).))))... ( -43.00) >DroEre_CAF1 25497 101 - 1 CCCAGUGCUUGAAGGGAAACCGUCG--AGAAUUCUGGAGAGGGUAGUUGAGUACUUAAGUGCCCGUUCUGCGGGAG-UUUUGGGCGACUUUGGGGCUCUGGCGG .((((..(((((.((....)).)))--))....))))...(((((.((((....)))).)))))...((((.((((-(((..((....))..))))))).)))) ( -44.20) >DroYak_CAF1 25431 101 - 1 CCCAGUGUUUGAAGGGAAACCGUGG--AGAGCUCUGGAGAGGGUAGUUGUGUACUUAAGUGCCCGUUCUGCGGCAG-UUUUGGGUGACUUUGGGGCUCUGGCGG (((((.((((.((((...(((....--(((((((((.((((((((.(((......))).)))))..))).))).))-)))).)))..)))).)))).)))).). ( -36.30) >consensus CCCAGUGCUUGAAGGGAAACCGUCC__AGAACUCUGGAGAGGGUAGUUGUGUACUUAAGUGCCCGUUCUGCGGGGG_UUUUGGGUGACUUUGGGGUUCUGGUGG .((((.((((.((((....(((.........(((((.((((((((.(((......))).)))))..))).))))).....)))....)))).)))).))))... (-23.91 = -24.37 + 0.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:00 2006