
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,948,837 – 1,948,933 |
| Length | 96 |
| Max. P | 0.944998 |

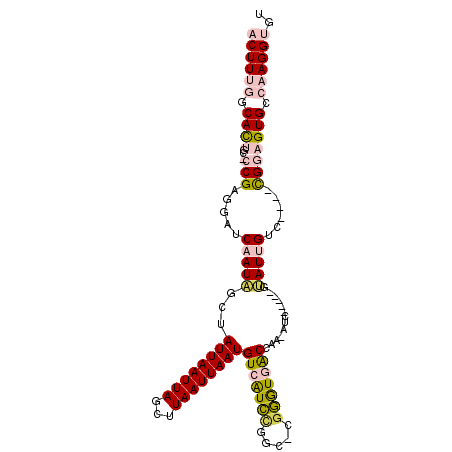
| Location | 1,948,837 – 1,948,933 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
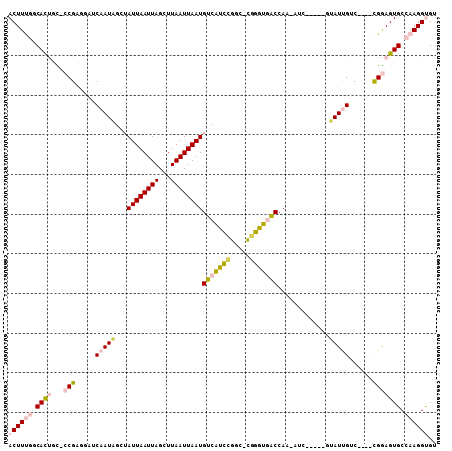
>2L_DroMel_CAF1 1948837 96 + 22407834 ACUUUGCCACUGU-CCGAGGAUCAAUAGCUAUUAAUUAGCUUAAUUAAUGUCGUCCGGC-CAGGUGACCAA-AUC-----GUAUUGUC----CGGAGUGCCAAGGUGU ((((((.(((..(-((..(((.(((((((((.....)))).........((((.((...-..))))))...-...-----.)))))))----))))))).)))))).. ( -26.10) >DroPse_CAF1 148223 105 + 1 ACUUC--CAUUUUGACGGCUAUCAAUUGAUAUUAAUUAGCUUAAUUAAUGUCAUUUGGC-UGAGUUGCCAAAAGCGGAUGGAAUCGACUCAAUGAAGUGCCAAGGCGU (((((--....((((((.(((((...((((((((((((...))))))))))))((((((-......))))))....)))))...))..)))).)))))((....)).. ( -29.80) >DroSec_CAF1 113531 97 + 1 ACUUUGGCACCGC-CCGAGGAUCAAUAGCUAUUAAUUAGCUUAAUUAAUGUCGUCCGGCCCGGGUGACCAA-GUC-----GUAUUGUC----CGGAGUGCCAAGGUGU ((((((((((.((-(.((.(((....(((((.....)))))........))).)).)))(((((..(....-...-----...)..))----))).)))))))))).. ( -37.10) >DroSim_CAF1 97518 96 + 1 ACUUUGGCACCGC-CCGAGGAUCAAUAGCUAUUAAUUAGCUUAAUUAAUGUCGUCCGGC-CGGGUGACCAA-GUC-----GUAUUGUC----CGGAGUGCCAAGGUGU ((((((((((..(-(...(((.(((((((((.....))))...............((((-.((....))..-)))-----))))))))----))).)))))))))).. ( -34.80) >DroYak_CAF1 124702 96 + 1 ACUUUGGCACUGC-CCGGGUAUCAAUAGGUAUUAAUUAGCUUAAUUAAUGUCAUCCGGC-CGGAUGACCAG-AUG-----UGAUUGUC----CGGAGUGCCAAGGUGU (((((((((((..-((((..((((.(((((........)))))......(((((((...-.)))))))...-...-----))).)..)----)))))))))))))).. ( -39.50) >DroPer_CAF1 146907 105 + 1 ACUUC--CAUUUUGACGGCUAUCAAUUGAUAUUAAUUAGCUUAAUUAAUGUCAUUUGGC-UGAGUUGCCAAAAGCGGAUGGAAUCGACUCAAUGAAGUGCCAAGGCGU (((((--....((((((.(((((...((((((((((((...))))))))))))((((((-......))))))....)))))...))..)))).)))))((....)).. ( -29.80) >consensus ACUUUGGCACUGC_CCGAGGAUCAAUAGCUAUUAAUUAGCUUAAUUAAUGUCAUCCGGC_CGGGUGACCAA_AUC_____GUAUUGUC____CGGAGUGCCAAGGUGU ((((((.((((...(((.....(((((...((((((((...))))))))(((((((.....))))))).............)))))......))))))).)))))).. (-14.00 = -14.67 + 0.67)

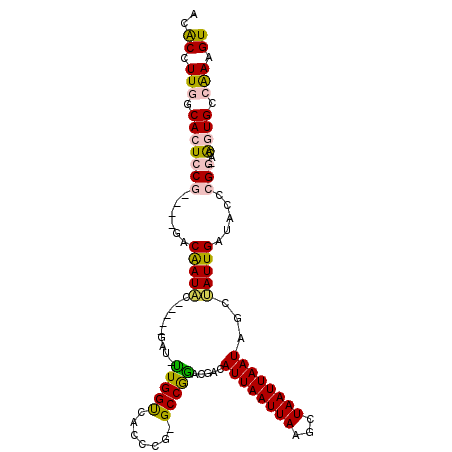
| Location | 1,948,837 – 1,948,933 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -11.14 |
| Energy contribution | -10.62 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
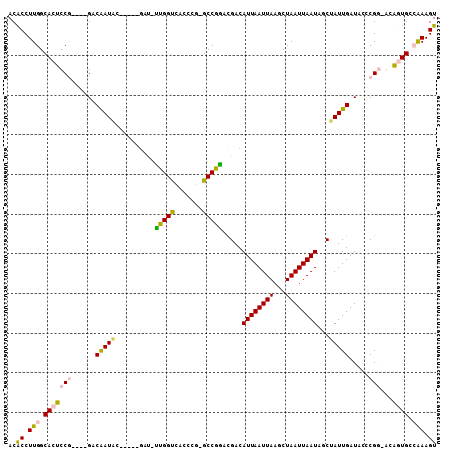
>2L_DroMel_CAF1 1948837 96 - 22407834 ACACCUUGGCACUCCG----GACAAUAC-----GAU-UUGGUCACCUG-GCCGGACGACAUUAAUUAAGCUAAUUAAUAGCUAUUGAUCCUCGG-ACAGUGGCAAAGU ..((.(((.(((((((----((((((.(-----(.(-((((((....)-))))))))..........(((((.....))))))))).)))..))-)..))).))).)) ( -27.10) >DroPse_CAF1 148223 105 - 1 ACGCCUUGGCACUUCAUUGAGUCGAUUCCAUCCGCUUUUGGCAACUCA-GCCAAAUGACAUUAAUUAAGCUAAUUAAUAUCAAUUGAUAGCCGUCAAAAUG--GAAGU .....(((((.(......).)))))((((((.....((((((......-))))))((((.(((((((...)))))))((((....))))...))))..)))--))).. ( -22.70) >DroSec_CAF1 113531 97 - 1 ACACCUUGGCACUCCG----GACAAUAC-----GAC-UUGGUCACCCGGGCCGGACGACAUUAAUUAAGCUAAUUAAUAGCUAUUGAUCCUCGG-GCGGUGCCAAAGU ..((.(((((((((((----(.(((...-----...-))).....))))(((.((.((..(((((..(((((.....)))))))))))).)).)-)))))))))).)) ( -31.50) >DroSim_CAF1 97518 96 - 1 ACACCUUGGCACUCCG----GACAAUAC-----GAC-UUGGUCACCCG-GCCGGACGACAUUAAUUAAGCUAAUUAAUAGCUAUUGAUCCUCGG-GCGGUGCCAAAGU ..((.(((((((((((----((((((.(-----(.(-(.((((....)-))))).))..........(((((.....))))))))).)))...)-).)))))))).)) ( -29.00) >DroYak_CAF1 124702 96 - 1 ACACCUUGGCACUCCG----GACAAUCA-----CAU-CUGGUCAUCCG-GCCGGAUGACAUUAAUUAAGCUAAUUAAUACCUAUUGAUACCCGG-GCAGUGCCAAAGU ..((.(((((((((((----(...((((-----(((-((((((....)-))))))))..((((((((...))))))))......))))..))))-..)))))))).)) ( -36.60) >DroPer_CAF1 146907 105 - 1 ACGCCUUGGCACUUCAUUGAGUCGAUUCCAUCCGCUUUUGGCAACUCA-GCCAAAUGACAUUAAUUAAGCUAAUUAAUAUCAAUUGAUAGCCGUCAAAAUG--GAAGU .....(((((.(......).)))))((((((.....((((((......-))))))((((.(((((((...)))))))((((....))))...))))..)))--))).. ( -22.70) >consensus ACACCUUGGCACUCCG____GACAAUAC_____GAU_UUGGUCACCCG_GCCGGACGACAUUAAUUAAGCUAAUUAAUAGCUAUUGAUACCCGG_ACAGUGCCAAAGU ..((.(((.(((((((......(((((..........(((((.......))))).....((((((((...))))))))...))))).....)))...)))).))).)) (-11.14 = -10.62 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:49 2006