
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,151,047 – 18,151,207 |
| Length | 160 |
| Max. P | 0.888355 |

| Location | 18,151,047 – 18,151,167 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -22.60 |
| Energy contribution | -21.47 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
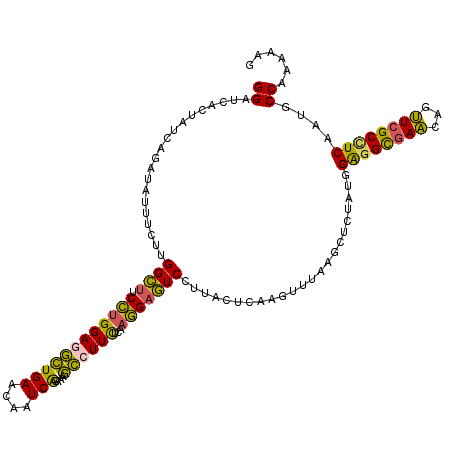
>2L_DroMel_CAF1 18151047 120 + 22407834 GGAUCACUAUCAGAUAUUUGUGGGCUUCCUGGAAGCUGAACAAUCAGAAAGCCUUUUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCAAAAAG ((.............(((((.(((((((.((((((((((....))))......)))))).))))))).....)))))............(((((((.....)))))))....))...... ( -30.20) >DroSec_CAF1 1438 120 + 1 GGAUCACUAUCAGAUAUUUGUUGGCUACCUGGAGGCUGAACAAUCAGAAAGCCUUCUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCCAAAAG ................((((..(((.....(((((((............))))))).((.(((((....((....))....))))).))(((((((.....)))))))...))))))).. ( -32.80) >DroSim_CAF1 1987 120 + 1 GGAUCACUAUCAGAUAUUUGUUGGCUACCUGGAGGCUGAACAAUCAGAAAGCCUUCUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCCAAAAG ................((((..(((.....(((((((............))))))).((.(((((....((....))....))))).))(((((((.....)))))))...))))))).. ( -32.80) >DroEre_CAF1 2573 120 + 1 GGAUCAUUAUCAGAUAUUUCUUGGCGUCCUGGAGGCUGAAAAAUCAGAACGCCUUCUCAAGGAGUCCUUACUGAAACUUAACCUCUGUGGAGGCGAACAAUUCGCUUCAAUGCCAAAAAG ....................(((((((...(((((.(((....((((((.(.((((....)))).).)).))))...))).)))))...(((((((.....))))))).))))))).... ( -30.90) >DroYak_CAF1 1791 120 + 1 GGAUCACUAUCAGAUAUUUCUUGGCUUCCUGGAGGCUGAAGAGUCGGAACGCCUUCUCAAGGAGUCCUUACUGAAAUUUAGCCUCUAUGGAGGCGAACAAUUCGCCUCAAUGCCAAAAAG ....................(((((..(.(((((((((((...((((((.(.((((....)))).).)).))))..))))))))))).)(((((((.....)))))))...))))).... ( -42.40) >DroAna_CAF1 22373 120 + 1 GGAUCGGUAUCAAAUAUUCCUUGGUUGCAAGGAAAGUGACAAGUCGGAGACUCUUCUCUGUGAAUCUCUGAAUCAGUUGAAAUUGUUUGGGGGUCAGCAGCUUGCAUCCAUGCCACAGCG .....(((((......(((((((....))))))).(((.(((((.(((((....)))))((..(((((..((.((((....))))))..)))))..)).))))))))..)))))...... ( -34.30) >consensus GGAUCACUAUCAGAUAUUUCUUGGCUUCCUGGAGGCUGAACAAUCAGAAAGCCUUCUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCAAAAAG ((....................((((.((((((((((((....)))....))))))...))))))).......................((((((((...))))))))....))...... (-22.60 = -21.47 + -1.13)

| Location | 18,151,087 – 18,151,207 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -20.71 |
| Energy contribution | -21.19 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18151087 120 + 22407834 CAAUCAGAAAGCCUUUUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCAAAAAGGUAUUUCAAUGUUGCCAACGACAUUAUUCAAGCGAAGAAU ...((.(((((((((((((.(((((....((....))....))))).))(((((((.....))))))).......))))))).))))(((((((....)))))))........))..... ( -32.80) >DroSec_CAF1 1478 120 + 1 CAAUCAGAAAGCCUUCUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCCAAAAGGUAUUUCAAUGUUGCCAACGACAUUAUUCAAGCGAAGAAU ...((.(((((((((..((.(((((....((....))....))))).))(((((((.....))))))).........))))).))))(((((((....)))))))........))..... ( -31.00) >DroSim_CAF1 2027 120 + 1 CAAUCAGAAAGCCUUCUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCCAAAAGGUAUUUCAAUGUUGCCAACGACAUUAUUCAAGCGAAGAAU ...((.(((((((((..((.(((((....((....))....))))).))(((((((.....))))))).........))))).))))(((((((....)))))))........))..... ( -31.00) >DroEre_CAF1 2613 120 + 1 AAAUCAGAACGCCUUCUCAAGGAGUCCUUACUGAAACUUAACCUCUGUGGAGGCGAACAAUUCGCUUCAAUGCCAAAAAGGUAUUUCAAUGUUGCCAACGACAUUAUUCAAGCGAAGAAC ...((....(((((((.((((((((..........)))...))).)).))))))).....(((((((.((((((.....))))))..(((((((....)))))))....))))))))).. ( -32.70) >DroYak_CAF1 1831 120 + 1 GAGUCGGAACGCCUUCUCAAGGAGUCCUUACUGAAAUUUAGCCUCUAUGGAGGCGAACAAUUCGCCUCAAUGCCAAAAAGGUAUUUCAAUGUGGCCAACGACAUUAUUCAAGCGAACAAU ..((((((...(((.....)))..))).............(((.(..(((((((((.....)))))))((((((.....)))))).))..).)))....))).................. ( -32.10) >DroAna_CAF1 22413 120 + 1 AAGUCGGAGACUCUUCUCUGUGAAUCUCUGAAUCAGUUGAAAUUGUUUGGGGGUCAGCAGCUUGCAUCCAUGCCACAGCGAUACGUUCAAGCUGCGACCAAAGUGAUCGAAGCCAAACAA ...(((((((.((........)).)))))))...........((((((((.((((.((((((((.((...(((....)))....)).))))))))))))...(....)....)))))))) ( -38.00) >consensus CAAUCAGAAAGCCUUCUCAAGGAGUCCUUACUCAAGUUUAAGCUCUAUGGAGGCGAACAGUUCGCCUCAAUGCCAAAAAGGUAUUUCAAUGUUGCCAACGACAUUAUUCAAGCGAAGAAU ..........((((((....((((..................))))..))))))......(((((...((((((.....))))))..(((((((....)))))))......))))).... (-20.71 = -21.19 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:47 2006