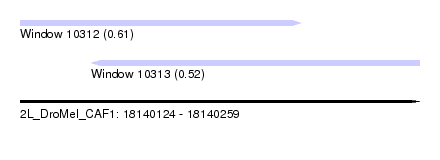
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,140,124 – 18,140,259 |
| Length | 135 |
| Max. P | 0.613410 |

| Location | 18,140,124 – 18,140,219 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.46 |
| Mean single sequence MFE | -20.67 |
| Consensus MFE | -5.12 |
| Energy contribution | -5.23 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
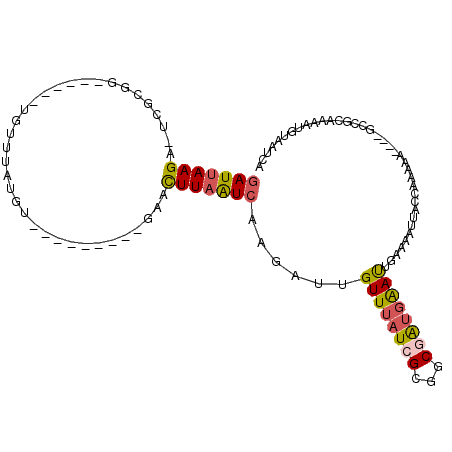
>2L_DroMel_CAF1 18140124 95 + 22407834 GAUUAAGA-UUGCGA------UGUUUAUGU---------GAACUUAGUCAAGAUUGUUUAUGGCGGCGAUGAAUUGAAAAUUACCAAAAACAAGGCCGCAAAAUGUAAUCA (((((((.-(..((.------......)).---------.).)))))))..(((((......(((((..((..(((........)))...))..)))))......))))). ( -22.20) >DroSec_CAF1 4314 95 + 1 GAUUAAGA-UCGCGG------UGUUUAUGU---------GAACUUAGUCAAGAUUGUUUAUCGCGGCGAUGAAUUGAAAAUUACCAAAAACAAGGCCGCAAAAUGUAAUCA (((((((.-(((((.------......)))---------)).)))))))..(((((......(((((..((..(((........)))...))..)))))......))))). ( -24.60) >DroSim_CAF1 4325 95 + 1 GAUUAAGA-UCGCGG------UGUUUAUGU---------GAACUUAGUCAAGAUUGUUUAUCGCGGCGAUGAAUUGAAAAUUACCAAAAACAAGGCCGCAAAAUGUAAUCA (((((((.-(((((.------......)))---------)).)))))))..(((((......(((((..((..(((........)))...))..)))))......))))). ( -24.60) >DroEre_CAF1 4300 72 + 1 GAUUAAGA-UCGCGG------UGUUUAUGU---------GAACUUAAUCAAGAUUGUUUAUCGCAUCGAUGGACUGAAAA-UACGAAAA---------------------- (((((((.-(((((.------......)))---------)).)))))))......((((((((...))))))))......-........---------------------- ( -18.40) >DroYak_CAF1 4344 73 + 1 GAUUAAGA-UCGCGG------UGUUUAUGU---------GAACUUAAUCAAGAUUGUUUAUCGCAGCGAUGAACUAAACAUUACCAAAA---------------------- (((((((.-(((((.------......)))---------)).)))))))......((((((((...))))))))...............---------------------- ( -18.80) >DroAna_CAF1 3890 107 + 1 CAUCAAGAAUUUCAACUUUUUGGCUUAUGAAAAAUUAGGGAGUUUCAUAAAUAUUGUUUAUCGCUUCAGCUAAUUAGGGAAUACUUCGAA----UCGGAGAAUUUUAAUCA ......((.......((..((((((..(....).....(((((...((((((...)))))).)))))))))))..))(((((.(((((..----.))))).)))))..)). ( -15.40) >consensus GAUUAAGA_UCGCGG______UGUUUAUGU_________GAACUUAAUCAAGAUUGUUUAUCGCGGCGAUGAAUUGAAAAUUACCAAAAA____GCCGCAAAAUGUAAUCA (((((((...................................)))))))......((((((((...))))))))..................................... ( -5.12 = -5.23 + 0.11)


| Location | 18,140,148 – 18,140,259 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -17.70 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 0.96 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18140148 111 - 22407834 CCUGGCCGUAGUGUACGUACAUACGUACACACAUACUCGUUGAUUACAUUUUGCGGCCUUGUUUUUGGUAAUUUUCAAUUCAUCGCCGCCAUAAACAAUCUUGACUAAGUU ...(((((((((((((((....))))))))........((.....))....)))))))((((((.((((..................)))).))))))............. ( -30.57) >DroSec_CAF1 4338 93 - 1 CCUGGCCGUAGUGUA------------------UACACGUUGAUUACAUUUUGCGGCCUUGUUUUUGGUAAUUUUCAAUUCAUCGCCGCGAUAAACAAUCUUGACUAAGUU ..........(((..------------------..)))(((((((.....(((((((..((...((((......))))..))..))))))).....))))..)))...... ( -20.20) >DroSim_CAF1 4349 93 - 1 CCUGGACGUAGUGUA------------------UACACGUUGAUUACAUUUUGCGGCCUUGUUUUUGGUAAUUUUCAAUUCAUCGCCGCGAUAAACAAUCUUGACUAAGUU ....(((((.((...------------------.)))))))((((.....(((((((..((...((((......))))..))..))))))).....))))........... ( -21.00) >consensus CCUGGCCGUAGUGUA__________________UACACGUUGAUUACAUUUUGCGGCCUUGUUUUUGGUAAUUUUCAAUUCAUCGCCGCGAUAAACAAUCUUGACUAAGUU ..........((((((((............))))))))(((((((.....(((((((..((...((((......))))..))..))))))).....))))..)))...... (-17.70 = -19.20 + 1.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:42 2006