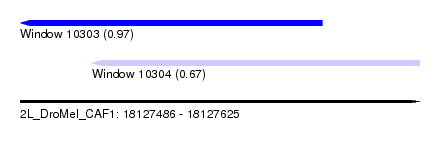
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,127,486 – 18,127,625 |
| Length | 139 |
| Max. P | 0.970471 |

| Location | 18,127,486 – 18,127,591 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.34 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -27.01 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18127486 105 - 22407834 CGUAUCUCCAAGUCCAGUUUGCAAUGGCUACUAGUAAACAAAUGGCUGCCGCUGUGGAUUGGCCAGGGACCCGAUUGCAAGGUAAAAAAC-AUUU---UCUCUUCUU---UU------ ((..((.((..(.((((((..((.((((..(((.........)))..)))).))..)))))))..))))..)).....(((..((((...-.)))---)..)))...---..------ ( -25.30) >DroVir_CAF1 20027 102 - 1 CGCAUCUCCAAGUCCAGUUUGCAGUGGCUGCUGGUCAACAAGUGGCUGCCGCUGUGGAUUGGCCAGGGACCCGAGUGCAAGGUGAG-CCUUAAAU---AAU-------AUUAA----- .((((.(((..(.((((((..(((((((.((((.(.....).)))).)))))))..)))))).)..))).....))))((((....-))))....---...-------.....----- ( -39.10) >DroPse_CAF1 20011 100 - 1 CGCAUAUCCAAGUCCAGUUUGCAGUGGCUGCUCGUGAACAAAUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAUUGCAAGGUAGG-AUUCA-----------ACUU---UAAAC--- .(((..(((..(.((((((..(((((((.((.(((......))))).)))))))..)))))).)..)))......)))(((((...-.....-----------))))---)....--- ( -37.80) >DroGri_CAF1 18955 114 - 1 CGCAUCUCCAAGUCCAGUUUGCAGUGGCUCCUGGUGAACAAGUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAGUGCAAGGUGAG-CAGCAGUU---AUUCAUCUGUAUUAAUCGAU ......(((..(.((((((..(((((((.((..(....)....))..)))))))..)))))).)..)))..((((((((.((((((-........---.)))))))))))...))).. ( -40.30) >DroYak_CAF1 31925 108 - 1 CGGAUCUCCAAGUCCAGUUUGCAAUGGCUGCUAGUAAACAAAUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAUUGCAAGGUAGA-AUCCCUUCGUUAUUUUUAUC---UU------ (((.((.((..(.((((((..((.((((.((((.........)))).)))).))..)))))))..)))).))).....((((((((-(............)))))))---))------ ( -36.10) >DroPer_CAF1 20039 100 - 1 CGCAUAUCCAAGUCCAGUUUGCAGUGGCAUCUCGUGAACAAAUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAUUGCAAGGUAGG-AUUCA-----------ACUU---UAAAC--- .(((..(((..(.((((((..((((((((.(.(((......)))).))))))))..)))))).)..)))......)))(((((...-.....-----------))))---)....--- ( -35.80) >consensus CGCAUCUCCAAGUCCAGUUUGCAGUGGCUGCUAGUGAACAAAUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAUUGCAAGGUAAG_AUUCA_UU___A_U__UCUU___UAA_____ ......(((..(.((((((..(((((((..(((.(.....).)))..)))))))..)))))).)..))).((........)).................................... (-27.01 = -27.35 + 0.34)

| Location | 18,127,511 – 18,127,625 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.15 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18127511 114 - 22407834 A---UCUGGAGAUACCAAUGGCAAU---GGGACUUUGAAUCGUAUCUCCAAGUCCAGUUUGCAAUGGCUACUAGUAAACAAAUGGCUGCCGCUGUGGAUUGGCCAGGGACCCGAUUGCAA .---..(((.....)))...(((((---(((((........))...(((..(.((((((..((.((((..(((.........)))..)))).))..)))))).)..)))))).))))).. ( -34.70) >DroVir_CAF1 20049 114 - 1 CUCCUCCUCGGAUGCC---GGCAAU---GGCACCUUGAAUCGCAUCUCCAAGUCCAGUUUGCAGUGGCUGCUGGUCAACAAGUGGCUGCCGCUGUGGAUUGGCCAGGGACCCGAGUGCAA ......(((((.((((---(....)---))))(((((....((........))((((((..(((((((.((((.(.....).)))).)))))))..)))))).)))))..)))))..... ( -46.20) >DroGri_CAF1 18989 117 - 1 UUCCUCGUCGGAUGGC---AGCAACAAUGGCACCUUGAAUCGCAUCUCCAAGUCCAGUUUGCAGUGGCUCCUGGUGAACAAGUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAGUGCAA ...((((..((.((.(---(.......)).))))............(((..(.((((((..(((((((.((..(....)....))..)))))))..)))))).)..)))..))))..... ( -40.10) >DroMoj_CAF1 22510 111 - 1 C---UCUUCGGAUUCC---GGCAAC---AAUACCUUGAACCGCAUAUCCAAGUCCAGUCUUCAGUGGCUGCUGGUCAACAAGUGGCUGCCUCUGUGGAUUGGCCAGGGACCCGAAUGCAA .---..(((((.((((---(((...---.....((((...........))))..((((((.(((.(((.((((.(.....).)))).))).))).))))))))).)))).)))))..... ( -37.60) >DroAna_CAF1 17802 108 - 1 G---UCGGGAGAUGCCA------AU---GGAACCUUGAACCGCAUCUCCAAGUCGAGCUUGCAGUGGCUGUUGGUGAACAAAUGGUUGCCAUUGUGGAUUGGCCAGGGUCCUGACUGCAA (---(((((((((((((------(.---(....))))....)))))(((..(((((..(..(((((((..(((.....)))......)))))))..).)))))..))))))))))..... ( -38.10) >DroPer_CAF1 20059 114 - 1 U---UCGGGAGAUGCCAAUGGGAAU---AACACUCUGAAUCGCAUAUCCAAGUCCAGUUUGCAGUGGCAUCUCGUGAACAAAUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAUUGCAA .---(((((..((((...(.(((..---.....))).)...)))).(((..(.((((((..((((((((.(.(((......)))).))))))))..)))))).)..))))))))...... ( -45.10) >consensus C___UCGGCAGAUGCC___GGCAAU___GGCACCUUGAAUCGCAUCUCCAAGUCCAGUUUGCAGUGGCUGCUGGUGAACAAAUGGCUGCCACUGUGGAUUGGCCAGGGACCCGAGUGCAA .........................................((((((((..(.((((((..(((((((..(((.(.....).)))..)))))))..)))))).)..)))...)).))).. (-25.62 = -26.15 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:34 2006