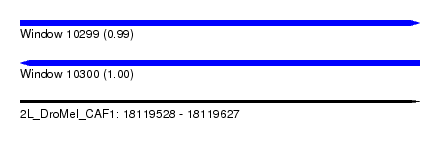
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,119,528 – 18,119,627 |
| Length | 99 |
| Max. P | 0.998325 |

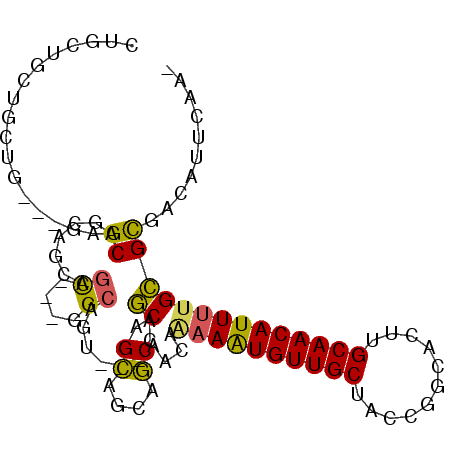
| Location | 18,119,528 – 18,119,627 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 71.31 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -18.13 |
| Energy contribution | -17.47 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
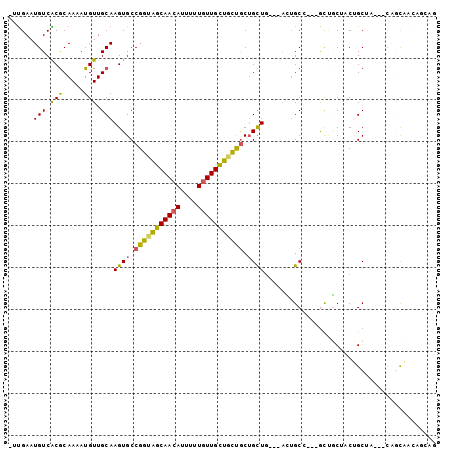
>2L_DroMel_CAF1 18119528 99 + 22407834 CUCGAAUGUCGCGCAAAAUGUUGCAAGUGCCGGUAGCAACACUUGUGUUGCUGUCGCUGCUGCCGAUUGCCCAUGUUGCUGCUGCCU---CAGCCACUGCCG ..........((((((....)))).((((..(..(((((((....)))))))..)((((..((.(...((.......))..).))..---)))))))))).. ( -28.90) >DroVir_CAF1 10729 89 + 1 -UUGAAUGUCACGCAAAAUGUUGCAAGUGCCGGCAGCAACAUUUUUGUUGCUGCUGCAGCUG---GCUGCG---ACUGCGACUGCGACGGCCGCAA------ -...........((....(((((((..(((.((((((((((....))))))))))((((...---.)))).---...)))..)))))))...))..------ ( -37.00) >DroPse_CAF1 10923 98 + 1 -UUGAAUGUCGCGCAAAAUGUUGCAAGUGCCGGUGGCAACAUUUCUGUUGCUGCUGCUGCUGCAGAGUGCCUCGGCCUCUCCAGCUC---CAGUUCCAGCAG -..((((((...((((....))))...((((...))))))))))((((((..((((..((((.((((.((....)))))).))))..---))))..)))))) ( -37.60) >DroGri_CAF1 9445 83 + 1 -UUGAAUGUCACACAAAAUGUUGCAAGUGCCGGGAGCAACAUUAUUGUUGUUGCUACUGUUG---GCAGCA---GCUGCAGCUGCUG------CAG------ -.....(((.(((.....))).))).....(((.(((((((.......))))))).)))...---((((((---((....)))))))------)..------ ( -30.00) >DroMoj_CAF1 13232 95 + 1 -UUGAAUGUCACGCAAAAUGUUGCAAGUGCCGGCAGCAACAUUUUUAUUGCUGCUGCUGCUA---ACUGCG---ACGGCAACUGCAACGGCAACAGCAACAG -...........((.....((((((..(((((((((((.((.......)).))))))(((..---...)))---.)))))..))))))(....).))..... ( -35.50) >DroPer_CAF1 10973 98 + 1 -UUGAAUGUCGCGCAAAAUGUUGCAAGUGCCGGUGGCAACAUUUCUGUUGCUGCUGCUGCUGCAGAGUGCCUCGGCCUCUCCAGCUC---CAGUUCCAGCAG -..((((((...((((....))))...((((...))))))))))((((((..((((..((((.((((.((....)))))).))))..---))))..)))))) ( -37.60) >consensus _UUGAAUGUCACGCAAAAUGUUGCAAGUGCCGGUAGCAACAUUUUUGUUGCUGCUGCUGCUG___ACUGCC___GCUGCUACUGCUA___CAGCAACAGCAG ......(((.(((.....))).)))((((.(((((((((((....))))))))))).))))......................................... (-18.13 = -17.47 + -0.66)

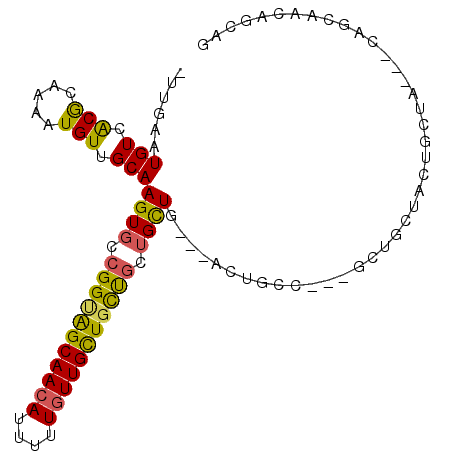
| Location | 18,119,528 – 18,119,627 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -13.93 |
| Energy contribution | -13.10 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.39 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18119528 99 - 22407834 CGGCAGUGGCUG---AGGCAGCAGCAACAUGGGCAAUCGGCAGCAGCGACAGCAACACAAGUGUUGCUACCGGCACUUGCAACAUUUUGCGCGACAUUCGAG (((..((.((((---..((....((.......)).....))..)))).))(((((((....))))))).)))...(.(((((....))))).)......... ( -29.30) >DroVir_CAF1 10729 89 - 1 ------UUGCGGCCGUCGCAGUCGCAGU---CGCAGC---CAGCUGCAGCAGCAACAAAAAUGUUGCUGCCGGCACUUGCAACAUUUUGCGUGACAUUCAA- ------.((((.....))))(((((.((---(((((.---...)))).(((((((((....))))))))).)))....((((....)))))))))......- ( -38.20) >DroPse_CAF1 10923 98 - 1 CUGCUGGAACUG---GAGCUGGAGAGGCCGAGGCACUCUGCAGCAGCAGCAGCAACAGAAAUGUUGCCACCGGCACUUGCAACAUUUUGCGCGACAUUCAA- .((((((..(((---..((((.((((((....)).)))).))))..)))..((((((....))))))..))))))(.(((((....))))).)........- ( -41.00) >DroGri_CAF1 9445 83 - 1 ------CUG------CAGCAGCUGCAGC---UGCUGC---CAACAGUAGCAACAACAAUAAUGUUGCUCCCGGCACUUGCAACAUUUUGUGUGACAUUCAA- ------..(------(((((((....))---))))))---.....((....(((.(((.((((((((...........)))))))))))))).))......- ( -28.00) >DroMoj_CAF1 13232 95 - 1 CUGUUGCUGUUGCCGUUGCAGUUGCCGU---CGCAGU---UAGCAGCAGCAGCAAUAAAAAUGUUGCUGCCGGCACUUGCAACAUUUUGCGUGACAUUCAA- .(((..(.((....(((((((.(((((.---.((...---..)).(((((((((.......)))))))))))))).))))))).....)))..))).....- ( -38.70) >DroPer_CAF1 10973 98 - 1 CUGCUGGAACUG---GAGCUGGAGAGGCCGAGGCACUCUGCAGCAGCAGCAGCAACAGAAAUGUUGCCACCGGCACUUGCAACAUUUUGCGCGACAUUCAA- .((((((..(((---..((((.((((((....)).)))).))))..)))..((((((....))))))..))))))(.(((((....))))).)........- ( -41.00) >consensus CUGCUGCUGCUG___GAGCAGCAGCAGC___GGCAGU___CAGCAGCAGCAGCAACAAAAAUGUUGCUACCGGCACUUGCAACAUUUUGCGCGACAUUCAA_ .................((.......((....))........((.((....))....((((((((((...........)))))))))))))).......... (-13.93 = -13.10 + -0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:30 2006