
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,107,751 – 18,107,919 |
| Length | 168 |
| Max. P | 0.918426 |

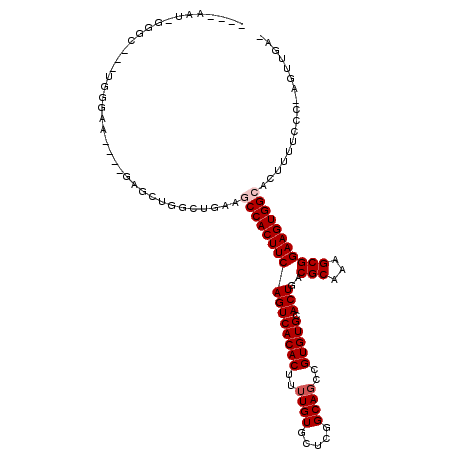
| Location | 18,107,751 – 18,107,856 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -24.57 |
| Energy contribution | -25.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
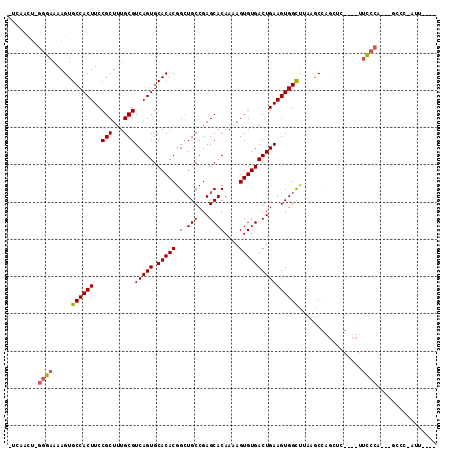
>2L_DroMel_CAF1 18107751 105 + 22407834 ----AAU-GGGC---UGGGAAAAUCGAGCUGGUUUCAGCCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC-AGUUGA- ----...-.(((---((((((((..(((.....))).((((((((((((((((..((((.....))))..))))).)))..(((...)))))))))))..)))))))-))))..- ( -42.10) >DroPse_CAF1 34969 101 + 1 UACCGAUGGGGA---UGGAA-----------GCAGAAACCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUUCCGAGCUGAA ....(((((((.---(((..-----------.......)))))))).))..........((((((.((..((((.((((..(((...)))..))))))))..)).)))))).... ( -30.80) >DroSec_CAF1 20657 105 + 1 ----AAU-GGGC---UGGGAAAAUCGAGCUGGUUUCAGCCACUUCAGUCACACUCUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC-AGUUGA- ----...-.(((---((((((((..(((.....))).((((((((((((((((..((((.....))))..))))).)))..(((...)))))))))))..)))))))-))))..- ( -42.10) >DroYak_CAF1 21008 105 + 1 ----AAU-GGGC---UGGGAAAAUCGAGCUGGCUUUAGCCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC-AGUUGA- ----...-.(((---((((((((..(((....)))..((((((((((((((((..((((.....))))..))))).)))..(((...)))))))))))..)))))))-))))..- ( -42.60) >DroAna_CAF1 26505 99 + 1 ----UGU-GGGUGGAUGGGGG----CAGGCGGCCGAGGCCACUUCAGUCACACUUUUGUGCUCGGCAUCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUGCA------GG- ----(((-(((((..((((((----((((.(((....))).)))..))).(((....))))))).)))))((((.((((..(((...)))..))))))))...)))------..- ( -35.50) >DroPer_CAF1 35207 101 + 1 UACCGAUGGGGC---UGGAA-----------GCAGAAACCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUUCCGAGCUGAA .........(((---(((((-----------(.((...(((((((((((((((..((((.....))))..))))).)))..(((...))))))))))..)).))))).))))... ( -33.90) >consensus ____AAU_GGGC___UGGGAA____GAGCUGGCUGAAGCCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC_AGUUGA_ .....................................((((((((((((((((..((((.....))))..))))).)))..(((...)))))))))))................. (-24.57 = -25.07 + 0.50)


| Location | 18,107,751 – 18,107,856 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.33 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
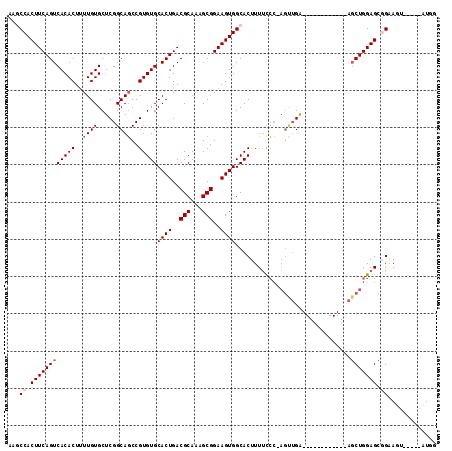
>2L_DroMel_CAF1 18107751 105 - 22407834 -UCAACU-GGGAAAAGUGCCACUUCCGCUUUGCGUCAGUGCACACGGCUGCCGAGCACAAAAGUGUGACUGAAGUGGCUGAAACCAGCUCGAUUUUCCCA---GCCC-AUU---- -....((-(((((((((((.(((..(((...)))..))))))).((((..(..((((((....)))).))...)..))))............))))))))---)...-...---- ( -36.90) >DroPse_CAF1 34969 101 - 1 UUCAGCUCGGAAAAAGUGCCACUUCCGCUUUGCGUCAGUGCACACGGCUGCCGAGCACAAAAGUGUGACUGAAGUGGUUUCUGC-----------UUCCA---UCCCCAUCGGUA ........((((..((.((((((..(((...)))(((((.(((((.(((....)))......))))))))))))))))..))..-----------)))).---............ ( -30.20) >DroSec_CAF1 20657 105 - 1 -UCAACU-GGGAAAAGUGCCACUUCCGCUUUGCGUCAGUGCACACGGCUGCCGAGCACAAGAGUGUGACUGAAGUGGCUGAAACCAGCUCGAUUUUCCCA---GCCC-AUU---- -....((-(((((((((((.(((..(((...)))..))))))).((((..(..((((((....)))).))...)..))))............))))))))---)...-...---- ( -36.90) >DroYak_CAF1 21008 105 - 1 -UCAACU-GGGAAAAGUGCCACUUCCGCUUUGCGUCAGUGCACACGGCUGCCGAGCACAAAAGUGUGACUGAAGUGGCUAAAGCCAGCUCGAUUUUCCCA---GCCC-AUU---- -....((-(((((((((((.(((..(((...)))..)))))))........((((((((....)))........((((....))))))))).))))))))---)...-...---- ( -38.10) >DroAna_CAF1 26505 99 - 1 -CC------UGCAAAGUGCCACUUCCGCUUUGCGUCAGUGCACACGGAUGCCGAGCACAAAAGUGUGACUGAAGUGGCCUCGGCCGCCUG----CCCCCAUCCACCC-ACA---- -..------.((((((((.......)))))))).(((((.(((((.(.(((...))))....)))))))))).(((((....)))))...----.............-...---- ( -32.30) >DroPer_CAF1 35207 101 - 1 UUCAGCUCGGAAAAAGUGCCACUUCCGCUUUGCGUCAGUGCACACGGCUGCCGAGCACAAAAGUGUGACUGAAGUGGUUUCUGC-----------UUCCA---GCCCCAUCGGUA ....(((.((((..((.((((((..(((...)))(((((.(((((.(((....)))......))))))))))))))))..))..-----------)))))---)).......... ( -33.00) >consensus _UCAACU_GGGAAAAGUGCCACUUCCGCUUUGCGUCAGUGCACACGGCUGCCGAGCACAAAAGUGUGACUGAAGUGGCUUAAGCCAGCUC____UUCCCA___GCCC_AUU____ ........((((.....((((((..(((...)))(((((.(((((.(.(((...))))....)))))))))))))))).................))))................ (-24.50 = -24.33 + -0.16)

| Location | 18,107,778 – 18,107,880 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18107778 102 + 22407834 CAGCCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC-AGUUGA------------AGCUGGAGCGGAAUUUCAGAAUGG ..((((((((((((((((..((((.....))))..))))).)))..(((...))))))))))).......((-(((...------------.))))).................. ( -33.80) >DroPse_CAF1 34990 107 + 1 AAACCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUUCCGAGCUGAAGCCCAACCUGAAGCUGGAGCUGGAG------CU-- ...(((((((((.....((((...((((((.((..((((.((((..(((...)))..))))))))..)).)))))).))))......)))))).)))(((....)------))-- ( -38.90) >DroSec_CAF1 20684 102 + 1 CAGCCACUUCAGUCACACUCUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC-AGUUGA------------AGCUGGAGCGGAAUUCCAGAAUGG ..((((((((((((((((..((((.....))))..))))).)))..(((...))))))))))).((.(((((-(((...------------.)))))...((....)).))).)) ( -36.20) >DroEre_CAF1 28726 102 + 1 UAGCCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC-AGUUGA------------AGCUGGAGCGGAAUUACAGAAUGG ..((((((((((((((((..((((.....))))..))))).)))..(((...))))))))))).......((-(((...------------.))))).................. ( -33.80) >DroAna_CAF1 26531 93 + 1 AGGCCACUUCAGUCACACUUUUGUGCUCGGCAUCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUGCA------GG------------GCCUGGAGCGGGAGCU----AUGU (((((...((((((((((....((((...))))..))))).))))).((((((.(.......).)))))).------.)------------))))..(((....)))----.... ( -34.60) >DroPer_CAF1 35228 107 + 1 AAACCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUUCCGAGCUGAAGCCCAACCUGAAGCUGGAGCUGGAG------CU-- ...(((((((((.....((((...((((((.((..((((.((((..(((...)))..))))))))..)).)))))).))))......)))))).)))(((....)------))-- ( -38.90) >consensus AAGCCACUUCAGUCACACUUUUGUGCUCGGCAGCCGUGUGCACUGACGCAAAGCGGAAGUGGCACUUUUCCC_AGUUGA____________AGCUGGAGCGGAAGU_____AUGG ...((.((((((((((((..((((.....))))..)))))((((..(((...)))..))))...............................))))))).))............. (-25.08 = -25.75 + 0.67)


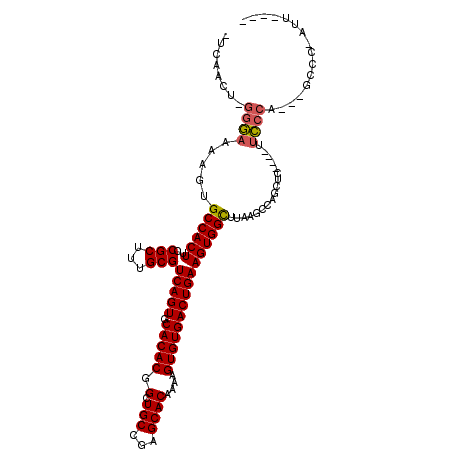
| Location | 18,107,818 – 18,107,919 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 84.26 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -24.64 |
| Energy contribution | -24.32 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18107818 101 + 22407834 CACUGACGCAAAGCGGAAGUGGCACUUUUCCCAGUUGAAGCUGGAGCGGAAUUUCAGAAUGGAUGGGGGGAUGGAAA-AUGGGAGCGGGGAAUAAGGAAUGG ((((..(((...)))..)))).((.((..((((.((.(..((((((.....))))))..).))))))..))))....-........................ ( -23.50) >DroSim_CAF1 20881 81 + 1 CACUGACGCAAAGCGGAAGUGGCACUUUUCCCAGUUGAAGCUGGAGCGGAAUUCCAGAAUGGAUGGGGGGAUGG---------------------GGAAUGA ((((..(((...)))..))))...((..((((.(((.(..((((((.....))))))..).)))...))))..)---------------------)...... ( -27.10) >DroEre_CAF1 28766 96 + 1 CACUGACGCAAAGCGGAAGUGGCACUUUUCCCAGUUGAAGCUGGAGCGGAAUUACAGAAUGGAUGGGGAGAUGGCGAGAU------GGGGAAUGGGGAAUGA ((((..(((...)))..))))((...((((((.(((.(..(((...........)))..).))).))))))..)).....------................ ( -24.80) >DroYak_CAF1 21075 95 + 1 CACUGACGCAAAGCGGAAGUGGCACUUUUCCCAGUUGAAGCUGGAGCGGAAUUCCAGAAUGGACGGGGAGAUGGGGA-AU------GAGGAAUGGAGAAUGG ((((..(((...)))..))))...(.((((((.(((.(..((((((.....))))))..).))).)))))).)....-..------................ ( -28.10) >consensus CACUGACGCAAAGCGGAAGUGGCACUUUUCCCAGUUGAAGCUGGAGCGGAAUUCCAGAAUGGAUGGGGAGAUGG_GA_AU______GGGGAAUGGGGAAUGA ((((..(((...)))..))))...(.((((((.(((.(..((((((.....))))))..).))).)))))).)............................. (-24.64 = -24.32 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:26 2006