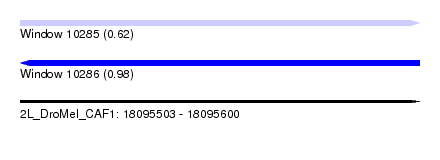
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,095,503 – 18,095,600 |
| Length | 97 |
| Max. P | 0.983374 |

| Location | 18,095,503 – 18,095,600 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -17.39 |
| Consensus MFE | -8.85 |
| Energy contribution | -9.68 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
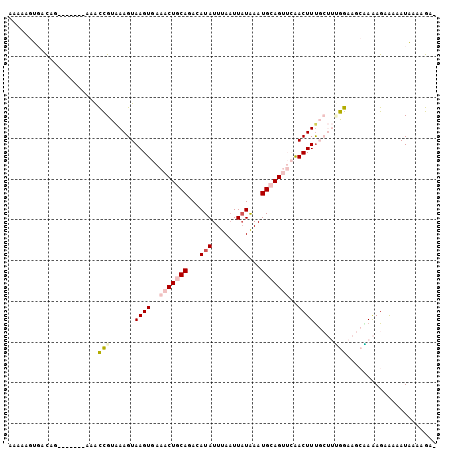
>2L_DroMel_CAF1 18095503 97 + 22407834 AAAAAGUGACUG-------AAAUCGUAAAGUAAGUGAAACUGCAGACAUAUUUAAUUAUAAAUGCAGUUCGACUUUGCUUUGGAAGCGAAAGAAAAAUAAAAGA- ............-------...(((((((((((((..(((((((...(((......)))...)))))))..)).)))))))....))))...............- ( -19.00) >DroPse_CAF1 21905 105 + 1 AAAAACCCAUAGAACCGUAGAGCUGUGAAGUAAGUAAAACUCCAGACAUAUUUAAUUAUGAAUGCAGCAAAACUUUCAAAGAGCAGACAGAGAGAAAUGGAAGAC ......((((...........((((((.(((.......))).)...((((......))))...)))))....(((((............)))))..))))..... ( -15.70) >DroSec_CAF1 8663 96 + 1 A-AAAGUGACUG-------AAACCGUAAAGUAAGUGAAACUGCAGACAUAUUUAUUUUUAAAUGCAGUUCGACUUUGCUUUGGAAGCAAAAGAAAAAUAAAAGA- .-...((..(..-------((...(((((((...((((.(((((..................))))))))))))))))))..)..)).................- ( -16.17) >DroSim_CAF1 8678 97 + 1 AAAAAGUGACUG-------AAACCGUAAAGUAAGUGAAACUGCAGACAUAUUUAAUUAUAAAUGCAGUUCGACUUUGCUUUGGAAGCAAAAGAAAAAUAAAAGA- .....((..(..-------((...(((((((...((((.(((((...(((......)))...))))))))))))))))))..)..)).................- ( -18.00) >DroEre_CAF1 16675 97 + 1 AAAAAGUGACAG-------AAGCCGUAAAGUAAGUGAAACUGCAGACAUAUUUAAUUAUAAAUGCAGUUCAACUUCGCUUCGGAAGCGAAAGAAAAAUAAAAGU- ............-------...(((..((((((((..(((((((...(((......)))...)))))))..)))).)))))))...(....)............- ( -19.80) >DroPer_CAF1 22052 105 + 1 AAAAACCCAUAGAACCGUAGAGCUGUGAAGUAAGUAAAACUCCAGACAUAUUUAAUUAUGAAUGCAGCAAAACUUUCAAAGAGCAGACAGAGAGAAAUGGAAGAC ......((((...........((((((.(((.......))).)...((((......))))...)))))....(((((............)))))..))))..... ( -15.70) >consensus AAAAAGUGACAG_______AAACCGUAAAGUAAGUGAAACUGCAGACAUAUUUAAUUAUAAAUGCAGUUCAACUUUGCUUUGGAAGCAAAAGAAAAAUAAAAGA_ ......................(((......((((..(((((((...(((......)))...)))))))..)))).....)))...................... ( -8.85 = -9.68 + 0.83)


| Location | 18,095,503 – 18,095,600 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -11.99 |
| Energy contribution | -12.72 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
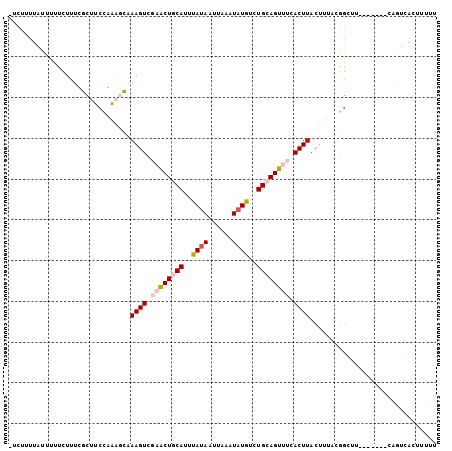
>2L_DroMel_CAF1 18095503 97 - 22407834 -UCUUUUAUUUUUCUUUCGCUUCCAAAGCAAAGUCGAACUGCAUUUAUAAUUAAAUAUGUCUGCAGUUUCACUUACUUUACGAUUU-------CAGUCACUUUUU -.................(((.....))).((((.((((((((..((((......))))..)))))))).)))).......(((..-------..)))....... ( -18.00) >DroPse_CAF1 21905 105 - 1 GUCUUCCAUUUCUCUCUGUCUGCUCUUUGAAAGUUUUGCUGCAUUCAUAAUUAAAUAUGUCUGGAGUUUUACUUACUUCACAGCUCUACGGUUCUAUGGGUUUUU ....(((((......((((..(((...(((((((...(((.((..((((......))))..)).)))...)))...)))).)))...))))....)))))..... ( -20.10) >DroSec_CAF1 8663 96 - 1 -UCUUUUAUUUUUCUUUUGCUUCCAAAGCAAAGUCGAACUGCAUUUAAAAAUAAAUAUGUCUGCAGUUUCACUUACUUUACGGUUU-------CAGUCACUUU-U -................((((.....))))((((.((((((((..((..........))..)))))))).))))............-------..........-. ( -14.30) >DroSim_CAF1 8678 97 - 1 -UCUUUUAUUUUUCUUUUGCUUCCAAAGCAAAGUCGAACUGCAUUUAUAAUUAAAUAUGUCUGCAGUUUCACUUACUUUACGGUUU-------CAGUCACUUUUU -................((((.....))))((((.((((((((..((((......))))..)))))))).))))............-------............ ( -17.80) >DroEre_CAF1 16675 97 - 1 -ACUUUUAUUUUUCUUUCGCUUCCGAAGCGAAGUUGAACUGCAUUUAUAAUUAAAUAUGUCUGCAGUUUCACUUACUUUACGGCUU-------CUGUCACUUUUU -..............((((((.....))))))((.((((((((..((((......))))..)))))))).)).........(((..-------..)))....... ( -21.70) >DroPer_CAF1 22052 105 - 1 GUCUUCCAUUUCUCUCUGUCUGCUCUUUGAAAGUUUUGCUGCAUUCAUAAUUAAAUAUGUCUGGAGUUUUACUUACUUCACAGCUCUACGGUUCUAUGGGUUUUU ....(((((......((((..(((...(((((((...(((.((..((((......))))..)).)))...)))...)))).)))...))))....)))))..... ( -20.10) >consensus _UCUUUUAUUUUUCUUUCGCUUCCAAAGCAAAGUCGAACUGCAUUUAUAAUUAAAUAUGUCUGCAGUUUCACUUACUUUACGGCUU_______CAGUCACUUUUU ..............................((((.((((((((..((((......))))..)))))))).))))............................... (-11.99 = -12.72 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:16 2006