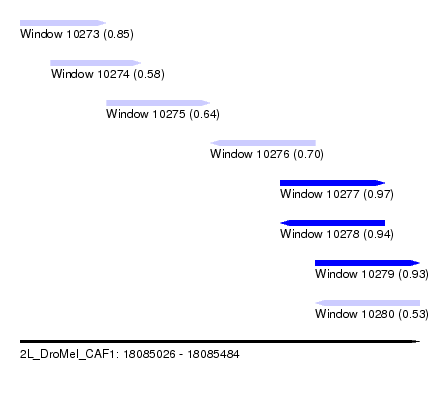
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,085,026 – 18,085,484 |
| Length | 458 |
| Max. P | 0.967786 |

| Location | 18,085,026 – 18,085,125 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -15.69 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085026 99 + 22407834 AAUUAAUAUAAAGUUUGUAAACUGAUAG---UAGAUGGGGGGUAAUUGAAAAAAACAGUAUGUAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA-AUAUAAA .............((((((..((.((..---...)).))..................((((((......))))))((((.((.....)).)))).-.)))))) ( -12.20) >DroSec_CAF1 292369 99 + 1 AAUUAAUAUAAAGUUUGUAAACUGAUAG---CAGAUGCGUGUUAAUGGA-AAAAACAGCAUGUAAUCAAAUAUGCGGCUACUAAACUAGAAGCCAAAUUUAUA .((((((((...((((((.........)---)))))..))))))))...-.......((((((......))))))((((.(((...))).))))......... ( -20.80) >DroSim_CAF1 294406 99 + 1 AAUUAAUAUAAAGUUUGUAAACUGAUAG---CAGAUGCGUGUUAAUUGAAAAAAACAGCAUGUAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA-AUUUAUA (((((((((...((((((.........)---)))))..)))))))))..........((((((......))))))((((.((.....)).)))).-....... ( -21.50) >DroYak_CAF1 309799 100 + 1 AAUUAAUAUAAAGUUUGUAAACUGAUAGCAGCAGAUGUGUGUUAAUUGAA--AAACAGCAUGCAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA-AUUUAUA ((((((((((..((((((............)))))).))))))))))...--.....(((((........)))))((((.((.....)).)))).-....... ( -19.60) >consensus AAUUAAUAUAAAGUUUGUAAACUGAUAG___CAGAUGCGUGUUAAUUGAAAAAAACAGCAUGUAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA_AUUUAUA (((((((((...(((((..............)))))..)))))))))..........((((((......))))))((((.((.....)).))))......... (-15.69 = -16.32 + 0.63)

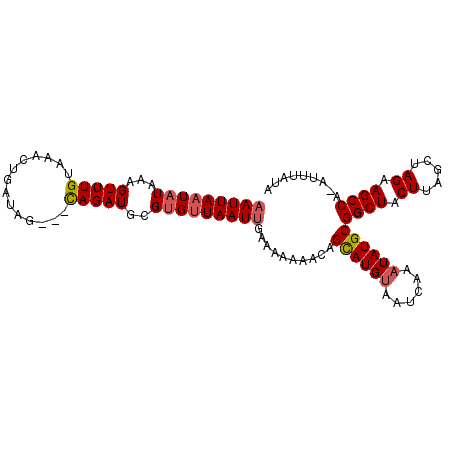
| Location | 18,085,061 – 18,085,165 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -12.79 |
| Energy contribution | -14.85 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085061 104 + 22407834 GGGGUAAUUGAAAAAAACAGUAUGUAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA-AUAUAAAAUGAAAUAUUGCAAUUAUCGCACUGCCUUCCGAAAAUUCC (.((((((((.........((((((......))))))((((.((.....)).)))).-..................)))))))).)................... ( -20.30) >DroSec_CAF1 292404 95 + 1 GUGUUAAUGGA-AAAAACAGCAUGUAAUCAAAUAUGCGGCUACUAAACUAGAAGCCAAAUUUAUAAUGAAAUAUC---------GCACUGCCUUCCGAAAAUUCU .......((((-(......((((((......))))))((((.(((...))).))))...................---------........)))))........ ( -17.30) >DroSim_CAF1 294441 104 + 1 GUGUUAAUUGAAAAAAACAGCAUGUAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA-AUUUAUAAUGAAAUAUCGCAGUUAUCGCACUGCCUUCCAAAAAUUCC (((.((((((.........((((((......))))))((((.((.....)).)))).-..................)))))).)))................... ( -20.60) >DroYak_CAF1 309837 101 + 1 GUGUUAAUUGAA--AAACAGCAUGCAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA-AUUUAUAAUGAAAUUUCGCAGUUAUU-CACUGCCUUCAAAAAAUGCC (((((..(((((--...(((((((........)))))((((.((.....)).)))).-........)).......(((((....-.))))).)))))..))))). ( -20.70) >consensus GUGUUAAUUGAAAAAAACAGCAUGUAAUCAAAUAUGCGGCUACUUAGCUAGAAGCCA_AUUUAUAAUGAAAUAUCGCAGUUAUCGCACUGCCUUCCAAAAAUUCC (((.((((((.........((((((......))))))((((.((.....)).))))....................)))))).)))................... (-12.79 = -14.85 + 2.06)

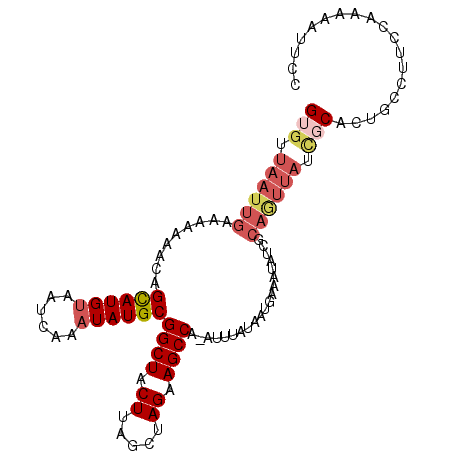
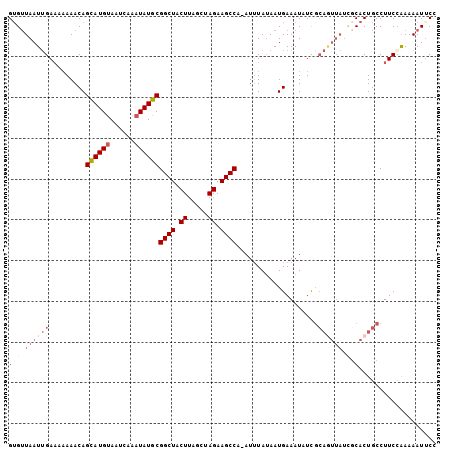
| Location | 18,085,125 – 18,085,244 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.05 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -19.83 |
| Energy contribution | -20.95 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085125 119 + 22407834 AUGAAAUAUUGCAAUUAUCGCACUGCCUUCCGAAAAUUCCGCA-AUUAAAACUGCGCCAGGCCAACAAUCAUGGCUUUUAGCAGCUGCAAAGUUGCAUUACGCAUACGCAGUGUAUGCCG ......((.(((((((...(((((((.............((((-........))))..((((((.......))))))...)))).)))..))))))).)).((((((.....)))))).. ( -31.80) >DroSec_CAF1 292468 111 + 1 AUGAAAUAUC---------GCACUGCCUUCCGAAAAUUCUGCAAAUUAAAACUGCGCCAGGCCAACAAUCAUGGCAUUUAGCAGCUGCAGAGUUGCAUUACGCAUACGCAGUGUAUGCCG ..........---------(((((((........(((((((((........((((.....((((.......)))).....)))).)))))))))((.....))....)))))))...... ( -32.30) >DroSim_CAF1 294505 119 + 1 AUGAAAUAUCGCAGUUAUCGCACUGCCUUCCAAAAAUUCCGCA-AUUAAAACUGCGCCAGGCCAACAAUCAUGGCAUUUAGCAGCUGCAGAGUUGCAUUACGCAUACGCAGUGUAUGCCG ..........(((((......)))))..............(((-(((....((((.....((((.......)))).....))))......)))))).....((((((.....)))))).. ( -30.10) >DroYak_CAF1 309899 117 + 1 AUGAAAUUUCGCAGUUAUU-CACUGCCUUCAAAAAAUGCCACA-AUUAAAACUACGCCUGGCCAACAAUCAUGGCUUUU-GCACUAACAAAGUUUCAGUACGCAUACGCGGUGUGUGCCG .((((((((.(((((....-.))))).................-...........((..(((((.......)))))...-)).......))))))))((((((((.....)))))))).. ( -28.00) >consensus AUGAAAUAUCGCAGUUAUCGCACUGCCUUCCAAAAAUUCCGCA_AUUAAAACUGCGCCAGGCCAACAAUCAUGGCAUUUAGCAGCUGCAAAGUUGCAUUACGCAUACGCAGUGUAUGCCG ...................(((((((..............(((.........)))((...((((.......)))).....((((((....)))))).....))....)))))))...... (-19.83 = -20.95 + 1.12)

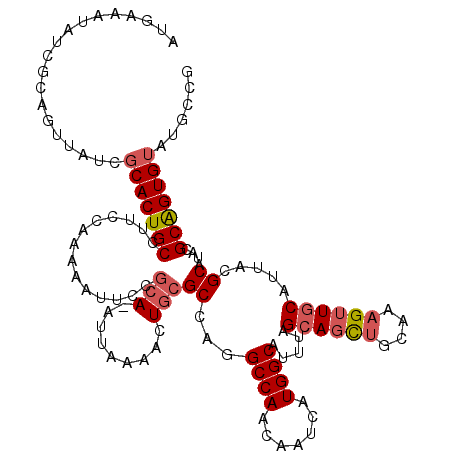
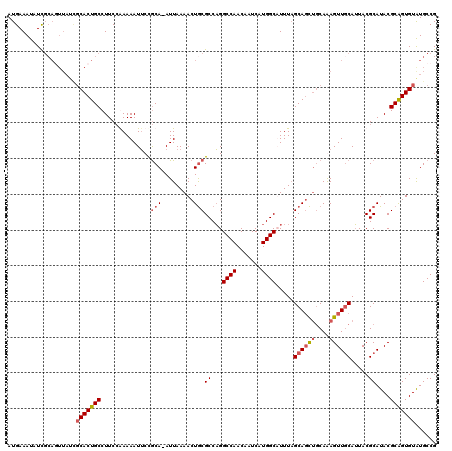
| Location | 18,085,244 – 18,085,364 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.28 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085244 120 - 22407834 AAAGAGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAGCCAGCUGGGGAAUGGAGCUCGCCUUUCAUAUGGCACUCGGGCUUAAUGGUCACUUCAUAAAUGCAGGCAAUUUGGGAGCU .....((...((((((((((((((..((((..(((......))).))))..)))).....(((........)))..))))))))))...)).((((.(((((.......))))))))).. ( -34.80) >DroSec_CAF1 292579 119 - 1 AAAGAGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAGCCAGCUGG-GAAUGGAGCUCUCAUUUCAUAUGGCACUCGGGCUUAAUGGUCACUUCAUAAAUGCAUGCAAUUUGGGAGCU .....((...(((((((((((((((.((((..(((......))).)))-)..(((((......)))))))))))...)))))))))...)).((((.(((((.......))))))))).. ( -31.00) >DroSim_CAF1 294624 120 - 1 AAAGAGGACUUUAAGCCUGGCCAUAACCCAAACUGAAAAGCCAGCUGGGGAAUGGAGCUUGCAUUUCAUAUGGCACUCGGGCUUAAUGGUCACUUCAUAAAUGCAGGCAAUUUGGGAGCU ......(((((((((((((((((((.((((..(((......))).))))(((((.......)))))..))))))...))))))))).)))).((((.(((((.......))))))))).. ( -36.70) >DroYak_CAF1 310016 120 - 1 GGAGUGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAGCCAGCUUGGGAAUGGAGCCGCCAUUUUAUAUGGCAGUCGGGCUUAAUGGUCACUUCAUAAAUGCAGGCGAUUUGGACGCU (((((((...((((((((((((((..(((((.(((......))).))))).))))....(((((.....)))))..))))))))))...))))))).........((((.......)))) ( -45.10) >consensus AAAGAGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAGCCAGCUGGGGAAUGGAGCUCGCAUUUCAUAUGGCACUCGGGCUUAAUGGUCACUUCAUAAAUGCAGGCAAUUUGGGAGCU .....((...(((((((((((((((.(((...(((......)))...)))..(((((......)))))))))))...)))))))))...)).((((.(((((.......))))))))).. (-28.52 = -28.28 + -0.25)

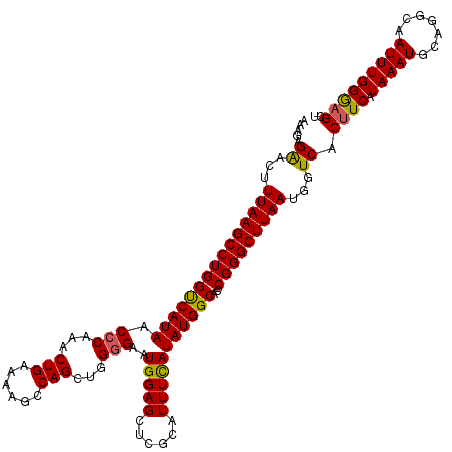
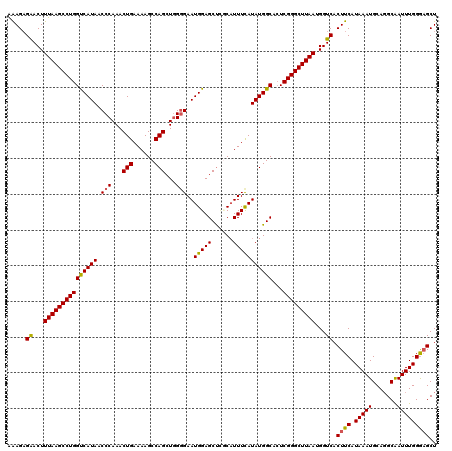
| Location | 18,085,324 – 18,085,444 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -38.21 |
| Energy contribution | -39.02 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085324 120 + 22407834 CUUUUCAGUUUGGGUUAUGACCAGGCUUAAAGUUCUCUUUUUAGCUGGCAGAAAGCCAGAGGCGAAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCUUUUGUCGACUGGU ....((((((((((((((..((.((((.((((....))))..))))(((.....)))...))(....)))))))))..((((((((.((((......)))).))))))))...)))))). ( -42.30) >DroSec_CAF1 292658 120 + 1 CUUUUCAGUUUGGGUUAUGACCAGGCUUAAAGUUCUCUUUUUAGCUGGCAGAAAGCCAGAGGCGAAAGGUGGCCCAAAAGAAGAUGGUCCUAGAUGCUGGAGCAUCUUUUGUCGACUGGU ....((((((((((((((..((.((((.((((....))))..))))(((.....)))...))(....)))))))))..((((((((.(((........))).))))))))...)))))). ( -41.60) >DroSim_CAF1 294704 120 + 1 CUUUUCAGUUUGGGUUAUGGCCAGGCUUAAAGUCCUCUUUUUAGCUGGCAGAAAGCCAUAGGCGAAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCUUUUGUCGACUGGU ....((((((((((((((.(((((.((.((((....))))..))))))).....(((...))).....))))))))..((((((((.((((......)))).))))))))...)))))). ( -44.50) >DroYak_CAF1 310096 119 + 1 CUUUUCAGUUUGGGUUAUGACCAGGCUUAAAGUUCACUCCUUAGCUGGCAGAAAGCCAGUGGCACAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCU-UUGUCGACUGGU ......(((((((.......)))))))...((((.(((((((.((((((.....))))))(((........)))..)))))(((((.((((......)))).)))))-..)).))))... ( -40.30) >consensus CUUUUCAGUUUGGGUUAUGACCAGGCUUAAAGUUCUCUUUUUAGCUGGCAGAAAGCCAGAGGCGAAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCUUUUGUCGACUGGU ....((((((((((((((..((.((((.((((....))))..))))(((.....)))...))(....)))))))))..((((((((.((((......)))).))))))))...)))))). (-38.21 = -39.02 + 0.81)

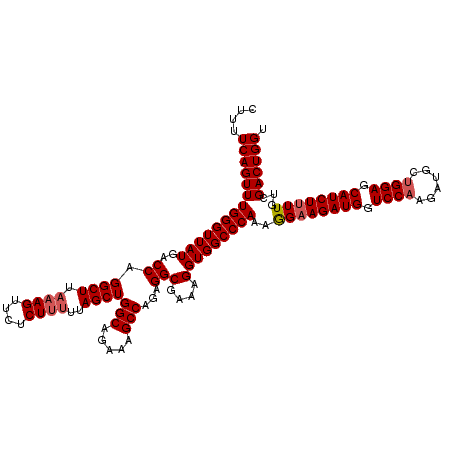
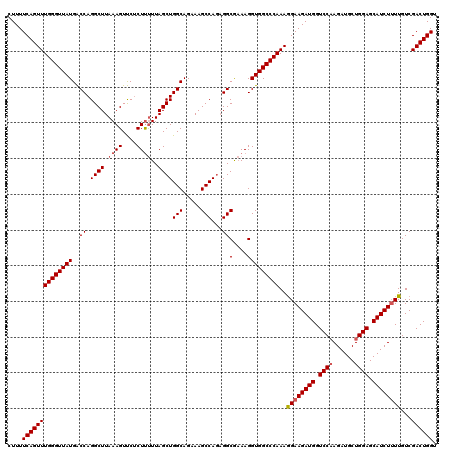
| Location | 18,085,324 – 18,085,444 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -29.44 |
| Energy contribution | -29.38 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085324 120 - 22407834 ACCAGUCGACAAAAGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUUCGCCUCUGGCUUUCUGCCAGCUAAAAAGAGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAG ..((((......(((((((....))))))).(((((((((..(((((((........))).(((((.....))))).))))..))))..........))))).........))))..... ( -30.70) >DroSec_CAF1 292658 120 - 1 ACCAGUCGACAAAAGAUGCUCCAGCAUCUAGGACCAUCUUCUUUUGGGCCACCUUUCGCCUCUGGCUUUCUGCCAGCUAAAAAGAGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAG ..((((.......((((((....))))))..((((((((.(((((((((........))))(((((.....)))))...))))))))..........))))).........))))..... ( -32.10) >DroSim_CAF1 294704 120 - 1 ACCAGUCGACAAAAGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUUCGCCUAUGGCUUUCUGCCAGCUAAAAAGAGGACUUUAAGCCUGGCCAUAACCCAAACUGAAAAG ..(((((((...((((((.(((((....))))).))))))...)))((((..((((......((((.....)))).....))))(((........))))))).........))))..... ( -31.90) >DroYak_CAF1 310096 119 - 1 ACCAGUCGACAA-AGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUGUGCCACUGGCUUUCUGCCAGCUAAGGAGUGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAG ..((((.....(-((((((....))))))).(((((((((((((.(((((.....).))).(((((.....)))))).)))))).))..........))))).........))))..... ( -35.60) >consensus ACCAGUCGACAAAAGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUUCGCCUCUGGCUUUCUGCCAGCUAAAAAGAGAACUUUAAGCCUGGUCAUAACCCAAACUGAAAAG ..((((......((((((.(((((....))))).))))))...(((((.........(((...))).....(((((((..((((....)))).)).))))).....)))))))))..... (-29.44 = -29.38 + -0.06)

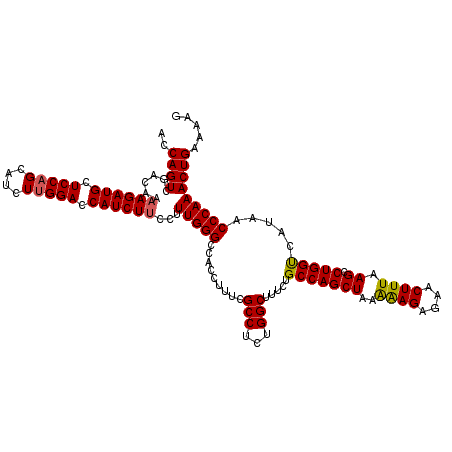
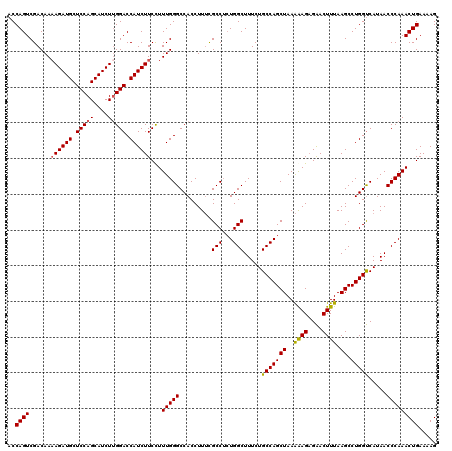
| Location | 18,085,364 – 18,085,484 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -35.67 |
| Energy contribution | -36.30 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085364 120 + 22407834 UUAGCUGGCAGAAAGCCAGAGGCGAAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCUUUUGUCGACUGGUCGGGUAAAAACAAUUUGCCCCAGGAUAGAAACGUUAAGCG (((((((((.....))))(.((((((..((.((((...((((((((.((((......)))).))))))))...((....))))))....))..)))))).)...........)))))... ( -39.80) >DroSec_CAF1 292698 120 + 1 UUAGCUGGCAGAAAGCCAGAGGCGAAAGGUGGCCCAAAAGAAGAUGGUCCUAGAUGCUGGAGCAUCUUUUGUCGACUGGUCGGGUAAAAACAAUUUGCCCCAGGAUAGAAACGUUAAGCG (((((((((.....))))(.((((((..((.((((...((((((((.(((........))).))))))))...((....))))))....))..)))))).)...........)))))... ( -39.10) >DroSim_CAF1 294744 120 + 1 UUAGCUGGCAGAAAGCCAUAGGCGAAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCUUUUGUCGACUGGUCGGGUAAAAACAAUUUGCCCCAGGAUAGAAACGUUAAGCG ...((((((.....)))...((((((..((.((((...((((((((.((((......)))).))))))))...((....))))))....))..)))))).................))). ( -39.20) >DroYak_CAF1 310136 118 + 1 UUAGCUGGCAGAAAGCCAGUGGCACAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCU-UUGUCGACUGGU-GGGUAAAAACAAUUUGCCCCAGGAUAGAAACGUUAAGCG ...((((((.....(((((((((....((....))....(((((((.((((......)))).)))))-))))).))))))-(((((((.....)))))))............))).))). ( -39.50) >consensus UUAGCUGGCAGAAAGCCAGAGGCGAAAGGUGGCCCAAAGGAAGAUGGUCCAAGAUGCUGGAGCAUCUUUUGUCGACUGGUCGGGUAAAAACAAUUUGCCCCAGGAUAGAAACGUUAAGCG ...((((((.....)))...((((((..((.((((...((((((((.((((......)))).))))))))...((....))))))....))..)))))).................))). (-35.67 = -36.30 + 0.63)

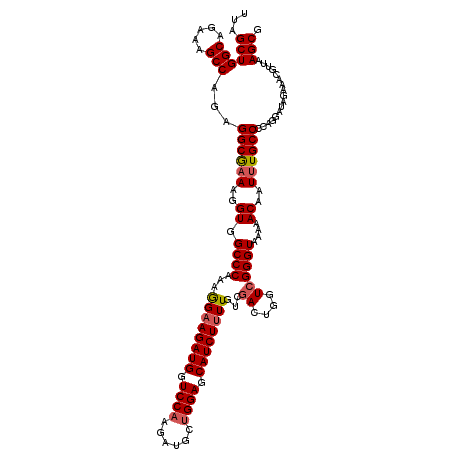
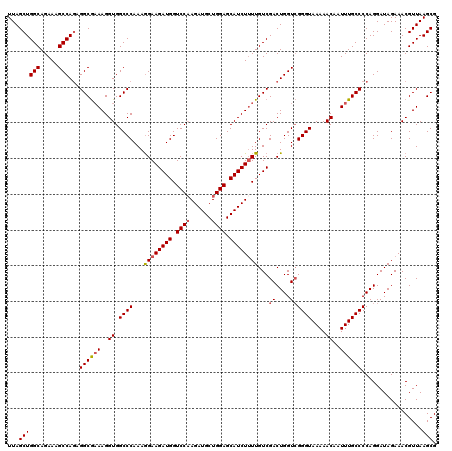
| Location | 18,085,364 – 18,085,484 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -26.95 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18085364 120 - 22407834 CGCUUAACGUUUCUAUCCUGGGGCAAAUUGUUUUUACCCGACCAGUCGACAAAAGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUUCGCCUCUGGCUUUCUGCCAGCUAA .((................(((((..............(((....)))....(((((((....)))))))((.(((........))).)).......)))))((((.....))))))... ( -32.00) >DroSec_CAF1 292698 120 - 1 CGCUUAACGUUUCUAUCCUGGGGCAAAUUGUUUUUACCCGACCAGUCGACAAAAGAUGCUCCAGCAUCUAGGACCAUCUUCUUUUGGGCCACCUUUCGCCUCUGGCUUUCUGCCAGCUAA .((................(((((..............(((....))).....((((((....)))))).((.(((........))).)).......)))))((((.....))))))... ( -31.30) >DroSim_CAF1 294744 120 - 1 CGCUUAACGUUUCUAUCCUGGGGCAAAUUGUUUUUACCCGACCAGUCGACAAAAGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUUCGCCUAUGGCUUUCUGCCAGCUAA .((.................((((..............(((....)))....(((((((....)))))))((.(((........))).)).......)))).((((.....))))))... ( -30.10) >DroYak_CAF1 310136 118 - 1 CGCUUAACGUUUCUAUCCUGGGGCAAAUUGUUUUUACCC-ACCAGUCGACAA-AGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUGUGCCACUGGCUUUCUGCCAGCUAA ..................(((.((((.............-...........(-((((((....)))))))((.(((........))).))...)))).)))(((((.....))))).... ( -30.10) >consensus CGCUUAACGUUUCUAUCCUGGGGCAAAUUGUUUUUACCCGACCAGUCGACAAAAGAUGCUCCAGCAUCUUGGACCAUCUUCCUUUGGGCCACCUUUCGCCUCUGGCUUUCUGCCAGCUAA .((................(((((..............(((....))).....((((((....)))))).((.(((........))).)).......)))))((((.....))))))... (-26.95 = -27.70 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:10 2006