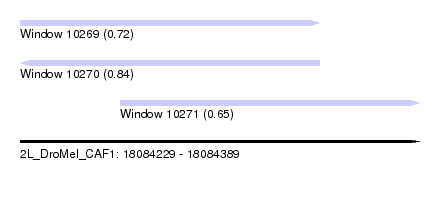
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,084,229 – 18,084,389 |
| Length | 160 |
| Max. P | 0.837044 |

| Location | 18,084,229 – 18,084,349 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.32 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
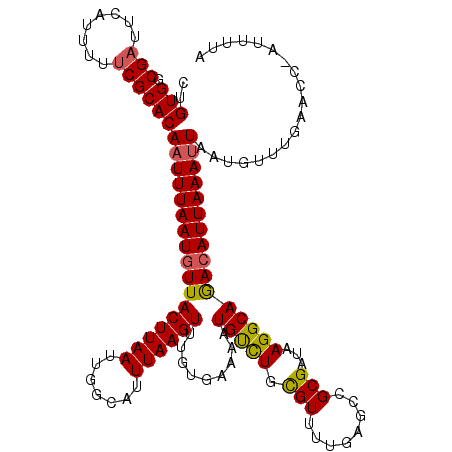
>2L_DroMel_CAF1 18084229 120 + 22407834 UUGUCAUUGUUGACAGUCUCUCUCUGCUCAAGGACGUGUUCUGUGCCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUUAAAAUGUCUGCGUUUUGAGCCG ((((((....)))))).........(((((((.((((....(((((.((........))))))).(((((((..((((((.......)))))).)))))))......))))))))))).. ( -30.00) >DroSec_CAF1 291585 120 + 1 UUGUCAUUGUUGACAGUCUCUCUCGGCUCAAGGACGUGUUCUGUGGCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUGAAAAUGUCUGUGUUUUGAGCCG ((((((....)))))).......((((((((((((((.(((....((((........))))((((...(((((((......))))))).....))))))).)))))).....)))))))) ( -35.10) >DroSim_CAF1 293608 120 + 1 UUGUCAUUGUUGACAGUCUCUCUCGGCUCAAGGACGUGUUCUGUGGCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUGAAAAUGUCUGUGUUUUGAGCCG ((((((....)))))).......((((((((((((((.(((....((((........))))((((...(((((((......))))))).....))))))).)))))).....)))))))) ( -35.10) >DroYak_CAF1 309003 118 + 1 UUUUCAUUGUUGACAGUCUCCCUCGGCUCAAGGACGUGUUCUGUGUCG-UUCGUUUUUCGCACAAUUUAAUUUUACUUAAUCGGCAUUUAAGUUUGUGUAAAUGCUUGCGUUUU-AGCAG .......((((((..........((((...((((....))))..))))-.........((((..(((((.....((((((.......)))))).....)))))...))))..))-)))). ( -21.00) >consensus UUGUCAUUGUUGACAGUCUCUCUCGGCUCAAGGACGUGUUCUGUGGCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUGAAAAUGUCUGCGUUUUGAGCCG ((((((....)))))).......(((((((((.((((....((((.(((........)))))))..................(((((((..........))))))).))))))))))))) (-24.70 = -25.32 + 0.62)

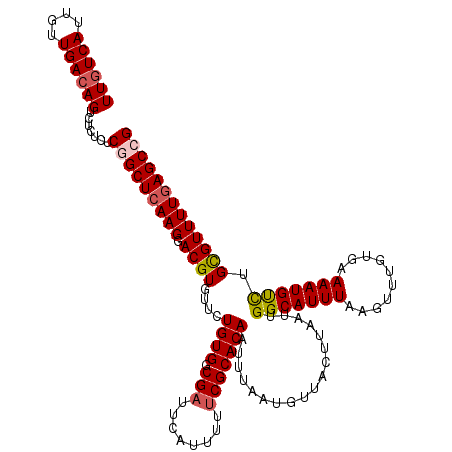
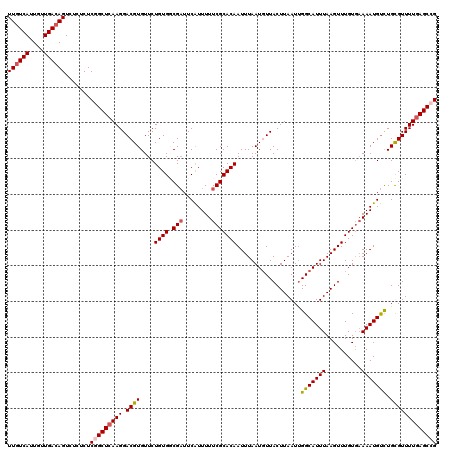
| Location | 18,084,229 – 18,084,349 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -20.02 |
| Energy contribution | -21.27 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18084229 120 - 22407834 CGGCUCAAAACGCAGACAUUUUAACAAACUUAAAUGCCAAUUAAGUAACAUUAAAUUGUGCGAAAAAUGAAUCGGCACAGAACACGUCCUUGAGCAGAGAGAGACUGUCAACAAUGACAA ..((((((.(((...............((((((.......)))))).........((((((((........)).))))))....)))..))))))..........(((((....))))). ( -23.20) >DroSec_CAF1 291585 120 - 1 CGGCUCAAAACACAGACAUUUUCACAAACUUAAAUGCCAAUUAAGUAACAUUAAAUUGUGCGAAAAAUGAAUCGCCACAGAACACGUCCUUGAGCCGAGAGAGACUGUCAACAAUGACAA ((((((((......(((..........((((((.......)))))).........((((((((........))).))))).....))).))))))))........(((((....))))). ( -27.20) >DroSim_CAF1 293608 120 - 1 CGGCUCAAAACACAGACAUUUUCACAAACUUAAAUGCCAAUUAAGUAACAUUAAAUUGUGCGAAAAAUGAAUCGCCACAGAACACGUCCUUGAGCCGAGAGAGACUGUCAACAAUGACAA ((((((((......(((..........((((((.......)))))).........((((((((........))).))))).....))).))))))))........(((((....))))). ( -27.20) >DroYak_CAF1 309003 118 - 1 CUGCU-AAAACGCAAGCAUUUACACAAACUUAAAUGCCGAUUAAGUAAAAUUAAAUUGUGCGAAAAACGAA-CGACACAGAACACGUCCUUGAGCCGAGGGAGACUGUCAACAAUGAAAA .(((.-.....))).(((((((........))))))).(((..(((.........(((((((.........-)).)))))....(..(((((...)))))..)))))))........... ( -19.10) >consensus CGGCUCAAAACACAGACAUUUUCACAAACUUAAAUGCCAAUUAAGUAACAUUAAAUUGUGCGAAAAAUGAAUCGCCACAGAACACGUCCUUGAGCCGAGAGAGACUGUCAACAAUGACAA ((((((((.((................((((((.......)))))).........((((((((........))).))))).....))..))))))))........(((((....))))). (-20.02 = -21.27 + 1.25)

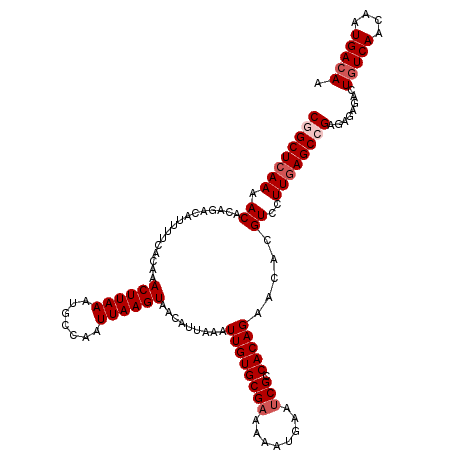
| Location | 18,084,269 – 18,084,389 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -20.91 |
| Energy contribution | -20.85 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18084269 120 + 22407834 CUGUGCCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUUAAAAUGUCUGCGUUUUGAGCCGCGAUAAGGCAGACAUUAAAUUAAUGUUUGAACCUACUUUA ..((((.((........))))))(((((((((((((((((.......))))))...((((((((....))))))))(((.......))).)))))))))))................... ( -26.80) >DroSec_CAF1 291625 119 + 1 CUGUGGCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUGAAAAUGUCUGUGUUUUGAGCCGCGAUAAGGCAGACAUUAAAUUAAUGUUUGAACC-AUUUUA .((((((.......((((((((.((((((((((((......))))).)))))))))))))))..((........))))))))....(((((((((((...)))))))))..))-...... ( -29.50) >DroSim_CAF1 293648 119 + 1 CUGUGGCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUGAAAAUGUCUGUGUUUUGAGCCGCGAUAAGGCAGACAUUAAAAUAAUGAUUGAAGC-AUUUUA .(((..((((.((((.((((((.((((((((((((......))))).)))))))))))))(((((((((.(((((.....))).)).))))))))).....))))))))..))-)..... ( -30.60) >DroYak_CAF1 309043 118 + 1 CUGUGUCG-UUCGUUUUUCGCACAAUUUAAUUUUACUUAAUCGGCAUUUAAGUUUGUGUAAAUGCUUGCGUUUU-AGCAGCGAUGAGGCAAACAUUAAAUUAAUAUUUUACCCUAUUUAG ..(((.((-.........)))))(((((((((((.(((.(((((((((((........)))))))((((.....-.)))))))).))).))).))))))))................... ( -20.90) >consensus CUGUGGCGAUUCAUUUUUCGCACAAUUUAAUGUUACUUAAUUGGCAUUUAAGUUUGUGAAAAUGUCUGCGUUUUGAGCCGCGAUAAGGCAGACAUUAAAUUAAUGUUUGAACC_AUUUUA ..(((.(((........))))))(((((((((((((((((.......)))))).........(((((.(((........)))...))))))))))))))))................... (-20.91 = -20.85 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:11:02 2006