
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,080,222 – 18,080,420 |
| Length | 198 |
| Max. P | 0.995020 |

| Location | 18,080,222 – 18,080,342 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -45.18 |
| Consensus MFE | -39.26 |
| Energy contribution | -40.08 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18080222 120 + 22407834 CCAGCAAAAGAUCUGGGUCUUCUCCAUGGGAGGCCUGAUCCUGGCAAUUGGGAUUUUUCUGGCCUCAGCCUGGCCAGCCGUAUCUCGGCAAUGGAUUCGAAGUCUUUUGCCCCUGGCUCC ((((((((((((..(((((((((....)))))))))(((((..((....(((((....((((((.......))))))....))))).))...)))))....)))))))))...))).... ( -46.40) >DroSec_CAF1 287438 120 + 1 CCAGCAAAAGAUCUGGGUCUUCUCCAUGGGAGGCCUGAUCCUGGCCAUUGGAAUUUUUCUGUCCUCAGCCUGGCCAGCCGUAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCC ((((((((((((..(((((((((....)))))))))(((((((((((..((.......(((....)))))))))))((((.....))))...)))))....)))))))))...))).... ( -44.21) >DroSim_CAF1 289482 120 + 1 CCAGCAAAAGAUCUGGGUUUUCUCCAUGGGAGGCCUGAUCCUGGCCAUUGGAAUUUUUCUGGCCUCUGCCUGGCCAGCCGUGUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCC ((((((((((((..(((((((((....)))))))))(((((((((((..(((.....)))(((....)))))))))((((.....))))...)))))....)))))))))...))).... ( -44.50) >DroYak_CAF1 304827 120 + 1 CCAGCAAAAGAUCUGGGUAUUCUCCAUGGGGGGCCUGAUCCUGGCGGUUGGGAUUUUUCUGGCCGCAGCCUGGCCCGCGGUAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCUUUGGCUCC (((((((((((.(((.(.....(((((...(((((.....(((.((((..(((...)))..)))))))...)))))((.(.....).)).)))))..).)))))))))))...))).... ( -45.60) >consensus CCAGCAAAAGAUCUGGGUCUUCUCCAUGGGAGGCCUGAUCCUGGCCAUUGGAAUUUUUCUGGCCUCAGCCUGGCCAGCCGUAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCC .........((((.(((((((((....)))))))))))))..(((((..(((((....((((((.......))))))....)))))(((((.((((.....)))).)))))..))))).. (-39.26 = -40.08 + 0.81)

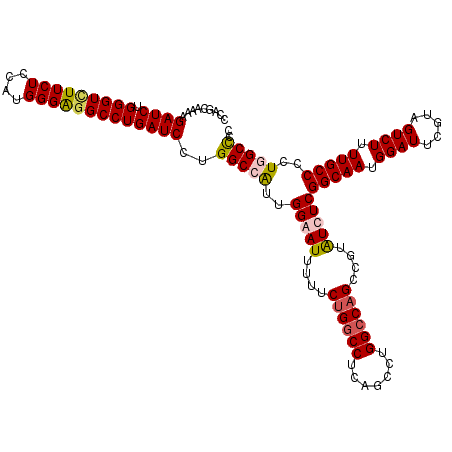
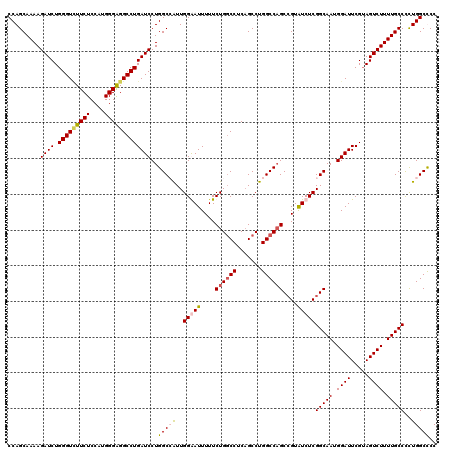
| Location | 18,080,262 – 18,080,381 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -29.02 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18080262 119 + 22407834 CUGGCAAUUGGGAUUUUUCUGGCCUCAGCCUGGCCAGCCGUAUCUCGGCAAUGGAUUCGAAGUCUUUUGCCCCUGGCUCCCAACUCUUUUAUCUACAAUCGCUGGGUGACUACG-CCCAC ..(((..((((((......(((((.......)))))((((......(((((.((((.....)))).)))))..)))))))))).................(.(((....)))))-))... ( -38.10) >DroSec_CAF1 287478 119 + 1 CUGGCCAUUGGAAUUUUUCUGUCCUCAGCCUGGCCAGCCGUAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCCCAACUCCUUUAUUUACAAUCGCUGGGUGACCACG-CCCAU ..(((.((((........(((....)))...((((((..(.....)(((((.((((.....)))).))))).)))))).................)))).)))(((((....))-))).. ( -33.40) >DroSim_CAF1 289522 120 + 1 CUGGCCAUUGGAAUUUUUCUGGCCUCUGCCUGGCCAGCCGUGUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCCCAACUCCUUUAUCUACAAUCGCUGGGGGACUACGGCCCAG ..((((....((((((...(((((.......)))))((((.....))))...))))))((((((....(((...)))(((((.(................).)))))))))))))))... ( -44.29) >DroYak_CAF1 304867 119 + 1 CUGGCGGUUGGGAUUUUUCUGGCCGCAGCCUGGCCCGCGGUAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCUUUGGCUCCCAACUCCUUUAUCUACAAUCGCUGGGUGACCACG-CCCAU ..(((((((((((........(((((.((...))..))))).....(((((.((((.....)))).)))))......)))))))..................(((....)))))-))... ( -42.00) >consensus CUGGCCAUUGGAAUUUUUCUGGCCUCAGCCUGGCCAGCCGUAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCCCAACUCCUUUAUCUACAAUCGCUGGGUGACCACG_CCCAU ..(((((..(((((....((((((.......))))))....)))))(((((.((((.....)))).)))))..)))))........................((((.........)))). (-29.02 = -29.90 + 0.88)

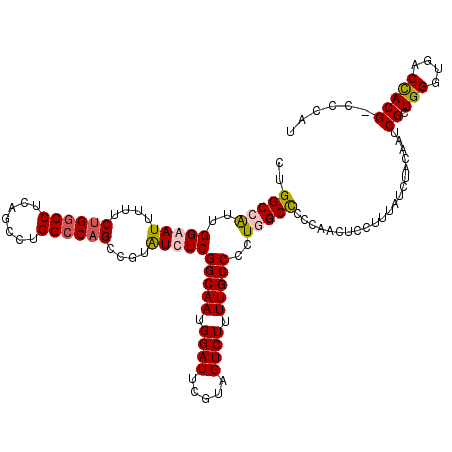

| Location | 18,080,262 – 18,080,381 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -36.16 |
| Energy contribution | -36.23 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18080262 119 - 22407834 GUGGG-CGUAGUCACCCAGCGAUUGUAGAUAAAAGAGUUGGGAGCCAGGGGCAAAAGACUUCGAAUCCAUUGCCGAGAUACGGCUGGCCAGGCUGAGGCCAGAAAAAUCCCAAUUGCCAG .((((-((.((((.((((((..((........))..)))))).(((...)))....)))).))........((((.....))))(((((.......))))).......))))........ ( -38.40) >DroSec_CAF1 287478 119 - 1 AUGGG-CGUGGUCACCCAGCGAUUGUAAAUAAAGGAGUUGGGGGCCAGGGGCAAAAGACUACGAAUCCAUUGCCGAGAUACGGCUGGCCAGGCUGAGGACAGAAAAAUUCCAAUGGCCAG ((((.-(((((((.((((((..((........))..)))))).(((...)))....)))))))...)))).((((.....))))((((((...((.(((........))))).)))))). ( -43.30) >DroSim_CAF1 289522 120 - 1 CUGGGCCGUAGUCCCCCAGCGAUUGUAGAUAAAGGAGUUGGGGGCCAGGGGCAAAAGACUACGAAUCCAUUGCCGAGACACGGCUGGCCAGGCAGAGGCCAGAAAAAUUCCAAUGGCCAG ((((..((((((((((((((..((........))..)))))))(((...)))....)))))))...(((((((((.....)))((((((.......))))))........)))))))))) ( -49.80) >DroYak_CAF1 304867 119 - 1 AUGGG-CGUGGUCACCCAGCGAUUGUAGAUAAAGGAGUUGGGAGCCAAAGGCAAAAGACUACGAAUCCAUUGCCGAGAUACCGCGGGCCAGGCUGCGGCCAGAAAAAUCCCAACCGCCAG .((((-(((((((.((((((..((........))..)))))).(((...)))....))))))).................((((((......))))))..........))))........ ( -38.50) >consensus AUGGG_CGUAGUCACCCAGCGAUUGUAGAUAAAGGAGUUGGGAGCCAGGGGCAAAAGACUACGAAUCCAUUGCCGAGAUACGGCUGGCCAGGCUGAGGCCAGAAAAAUCCCAAUGGCCAG .((((.(((((((.((((((..((........))..)))))).(((...)))....)))))))........((((.....)))).((((.......))))........))))........ (-36.16 = -36.23 + 0.06)

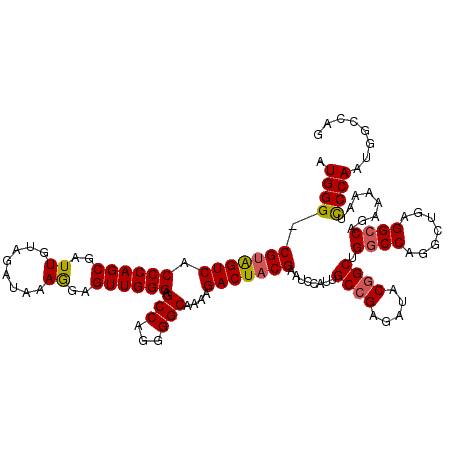
| Location | 18,080,302 – 18,080,420 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -31.31 |
| Consensus MFE | -25.60 |
| Energy contribution | -26.91 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18080302 118 + 22407834 UAUCUCGGCAAUGGAUUCGAAGUCUUUUGCCCCUGGCUCCCAACUCUUUUAUCUACAAUCGCUGGGUGACUACG-CCCACGCCGGUAUAU-AGCACAGUUUUCCUCUUCAACUGGACUAA (((..((((..(((((..((((......(((...)))........)))).))))).....(.((((((....))-)))))))))..)))(-((..(((((.........)))))..))). ( -29.74) >DroSec_CAF1 287518 118 + 1 UAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCCCAACUCCUUUAUUUACAAUCGCUGGGUGACCACG-CCCAUGCCGGUAUAU-AGCACAGUUUUCCUCUUCAACUGGACUAA (((..(((((.(((....(.(((.....(((...))).....))).).............(((((....))).)-)))))))))..)))(-((..(((((.........)))))..))). ( -29.40) >DroSim_CAF1 289562 120 + 1 UGUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCCCAACUCCUUUAUCUACAAUCGCUGGGGGACUACGGCCCAGGCCGGUAUAUGAGCACAGUUUUCCUUUUCAAAUGGACUAA (((((((((((.((((.....)))).)))))..(((((((((.(................).)))))).)))((((....))))......))).)))....(((.........))).... ( -35.49) >DroYak_CAF1 304907 118 + 1 UAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCUUUGGCUCCCAACUCCUUUAUCUACAAUCGCUGGGUGACCACG-CCCAUGCCGCUAUAU-AGCACAGUUUUCCUCUUUAACUGGACUAA (((..(((((.(((....((((..........((((...)))).........))))....(((((....))).)-)))))))))..)))(-((..(((((.........)))))..))). ( -30.61) >consensus UAUCUCGGCAAUGGAUUCGUAGUCUUUUGCCCCUGGCCCCCAACUCCUUUAUCUACAAUCGCUGGGUGACCACG_CCCAUGCCGGUAUAU_AGCACAGUUUUCCUCUUCAACUGGACUAA (((..(((((.(((...(((((((....(((...))).((((.(................).)))).)))))))..))))))))..)))..((..(((((.........)))))..)).. (-25.60 = -26.91 + 1.31)

| Location | 18,080,302 – 18,080,420 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -38.19 |
| Energy contribution | -38.38 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18080302 118 - 22407834 UUAGUCCAGUUGAAGAGGAAAACUGUGCU-AUAUACCGGCGUGGG-CGUAGUCACCCAGCGAUUGUAGAUAAAAGAGUUGGGAGCCAGGGGCAAAAGACUUCGAAUCCAUUGCCGAGAUA .(((..(((((.........)))))..))-).....((((((((.-((.((((.((((((..((........))..)))))).(((...)))....)))).))...))).)))))..... ( -39.70) >DroSec_CAF1 287518 118 - 1 UUAGUCCAGUUGAAGAGGAAAACUGUGCU-AUAUACCGGCAUGGG-CGUGGUCACCCAGCGAUUGUAAAUAAAGGAGUUGGGGGCCAGGGGCAAAAGACUACGAAUCCAUUGCCGAGAUA .(((..(((((.........)))))..))-).....((((((((.-(((((((.((((((..((........))..)))))).(((...)))....)))))))...))).)))))..... ( -44.10) >DroSim_CAF1 289562 120 - 1 UUAGUCCAUUUGAAAAGGAAAACUGUGCUCAUAUACCGGCCUGGGCCGUAGUCCCCCAGCGAUUGUAGAUAAAGGAGUUGGGGGCCAGGGGCAAAAGACUACGAAUCCAUUGCCGAGACA ...(((....(((..((.....))....))).....((((.((((.((((((((((((((..((........))..)))))))(((...)))....)))))))..))))..)))).))). ( -39.60) >DroYak_CAF1 304907 118 - 1 UUAGUCCAGUUAAAGAGGAAAACUGUGCU-AUAUAGCGGCAUGGG-CGUGGUCACCCAGCGAUUGUAGAUAAAGGAGUUGGGAGCCAAAGGCAAAAGACUACGAAUCCAUUGCCGAGAUA .(((..(((((.........)))))..))-)(((..((((((((.-(((((((.((((((..((........))..)))))).(((...)))....)))))))...))).)))))..))) ( -44.30) >consensus UUAGUCCAGUUGAAGAGGAAAACUGUGCU_AUAUACCGGCAUGGG_CGUAGUCACCCAGCGAUUGUAGAUAAAGGAGUUGGGAGCCAGGGGCAAAAGACUACGAAUCCAUUGCCGAGAUA ..((..(((((.........)))))..)).......((((((((..(((((((.((((((..((........))..)))))).(((...)))....)))))))...))).)))))..... (-38.19 = -38.38 + 0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:54 2006