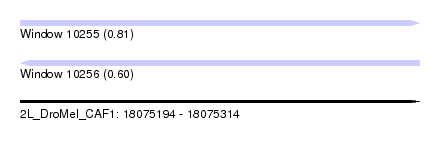
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,075,194 – 18,075,314 |
| Length | 120 |
| Max. P | 0.809692 |

| Location | 18,075,194 – 18,075,314 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -47.63 |
| Consensus MFE | -38.03 |
| Energy contribution | -37.95 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18075194 120 + 22407834 UGCGAUCGAUCGCAUCGCAGGAGCACCAUUAACCUGGGCAUCCGCAAGGUGCAGUACGCACCGACCAAGCAGGGCAUCCAGCCCUGCACCGUCGUUCGCAAGGACUUCCUUCUGUCGCCC (((((((....).))))))(((((.(((......))))).)))((..(((((.....)))))(((...(((((((.....)))))))......((((....))))........))))).. ( -50.80) >DroVir_CAF1 362054 120 + 1 UGCGAUCGUUCGCAUAGACGCAGCACUAUUAACCUGGGCAUACGCAAGGUGCAGUAUGCACCCACCAAACAGGGCAUGCAGCCCUGCACGGUGGUGCGCAAGGACUUUCUGUUGUCGCCC (((((....)))))..(((((.((((.........((((((((((.....)).)))))).))((((...((((((.....))))))...))))))))))...(((.....)))))).... ( -47.50) >DroGri_CAF1 375270 120 + 1 UGCGAUCGCUCGCAUCGUCGCAGCACCAUCAAUCUGGGCAUACGCAAGGUGCAAUAUGCACCCACCAAGGAUGGUAUGCAACCAUGCACGGUGGUGCGCAAGGACUUCCUUUUGUCGCCC .(((((.....((((.((.(((..((((((....(((((....))..(((((.....)))))..)))..)))))).))).)).))))..((.(((.(....).))).))....))))).. ( -46.50) >DroWil_CAF1 362641 120 + 1 UGCGAUCGUUCGCAUCGUCGCAGCACCAUCAAUUUGGGCAUCCGCAAGGUGCAGUACGCACCAACCAAACAGGGCAUUCAGCCCUGCACAGUGGUACGUAAGGAUUUCCUUUUGUCGCCC (((((.((.......))))))).............((((..((((..(((((.....))))).......((((((.....))))))....)))).(((.((((....)))).))).)))) ( -43.90) >DroMoj_CAF1 426678 120 + 1 UGCGAUCGCUCGCAUCGUCGCAGCACCAUCAAUCUGGGCAUACGCAAGGUGCAGUAUGCACCCACCAAGCAGGGCUUGCAGCCGUGCACAGUGGUGCGCAAGGACUUCCUGCUGUCGCCC .(((((.((..........((.(((((((.....(((((....))..(((((.....)))))..))).(((.(((.....))).)))...))))))))).(((....))))).))))).. ( -48.90) >DroAna_CAF1 266161 120 + 1 UGCGACCGGUCGCAUCGCCGCAGCACCAUCAACCUGGGCAUCCGCAAGGUGCAGUACGCGCCCACCAAACAGGGCAUCCAGCCCUGCACCGUCGUCCGCAAGGACUUCCUCUUGUCGCCC .(((((((((.(((..((.((....(((......)))(((.((....))))).....))((((........)))).....))..)))))))..((((....))))........))))).. ( -48.20) >consensus UGCGAUCGCUCGCAUCGUCGCAGCACCAUCAACCUGGGCAUACGCAAGGUGCAGUACGCACCCACCAAACAGGGCAUGCAGCCCUGCACAGUGGUGCGCAAGGACUUCCUUUUGUCGCCC (((((....))))).....................((((....((..(((((.....)))))(((((..((((((.....)))))).....))))).))...(((........))))))) (-38.03 = -37.95 + -0.08)

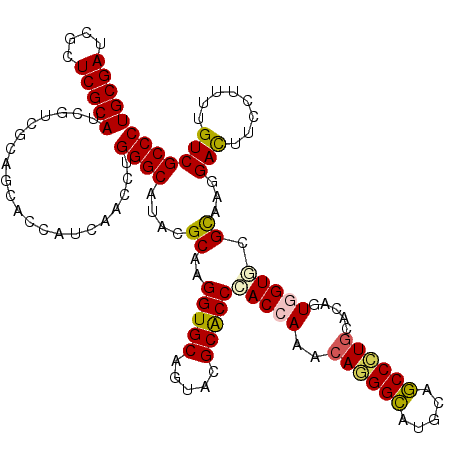
| Location | 18,075,194 – 18,075,314 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.33 |
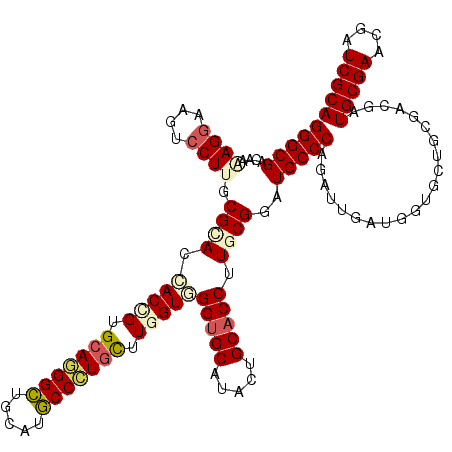
| Mean single sequence MFE | -51.90 |
| Consensus MFE | -41.62 |
| Energy contribution | -42.43 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18075194 120 - 22407834 GGGCGACAGAAGGAAGUCCUUGCGAACGACGGUGCAGGGCUGGAUGCCCUGCUUGGUCGGUGCGUACUGCACCUUGCGGAUGCCCAGGUUAAUGGUGCUCCUGCGAUGCGAUCGAUCGCA ..((((..((.((....))(((((...(((.(.(((((((.....))))))).).)))(((((.....)))))((((((..(((((......))).))..)))))))))))))..)))). ( -54.00) >DroVir_CAF1 362054 120 - 1 GGGCGACAACAGAAAGUCCUUGCGCACCACCGUGCAGGGCUGCAUGCCCUGUUUGGUGGGUGCAUACUGCACCUUGCGUAUGCCCAGGUUAAUAGUGCUGCGUCUAUGCGAACGAUCGCA (((((((........)))..((((((.(((((.(((((((.....))))))).)))))(((((.....))))).)))))).))))......((((........))))((((....)))). ( -53.60) >DroGri_CAF1 375270 120 - 1 GGGCGACAAAAGGAAGUCCUUGCGCACCACCGUGCAUGGUUGCAUACCAUCCUUGGUGGGUGCAUAUUGCACCUUGCGUAUGCCCAGAUUGAUGGUGCUGCGACGAUGCGAGCGAUCGCA (((((....((((....)))).))).))....(((((.(((((((((((((.((((.((((((.....)))))).((....))))))...))))))).)))))).))))).((....)). ( -50.30) >DroWil_CAF1 362641 120 - 1 GGGCGACAAAAGGAAAUCCUUACGUACCACUGUGCAGGGCUGAAUGCCCUGUUUGGUUGGUGCGUACUGCACCUUGCGGAUGCCCAAAUUGAUGGUGCUGCGACGAUGCGAACGAUCGCA (((((....((((....))))(((((((((((.(((((((.....))))))).))).)))))))).((((.....)))).))))).....(((.((..(((......))).)).)))... ( -51.20) >DroMoj_CAF1 426678 120 - 1 GGGCGACAGCAGGAAGUCCUUGCGCACCACUGUGCACGGCUGCAAGCCCUGCUUGGUGGGUGCAUACUGCACCUUGCGUAUGCCCAGAUUGAUGGUGCUGCGACGAUGCGAGCGAUCGCA ..((((..(((((....)))(((((.((((((.(((.(((.....))).))).)))))))))))...((((..(((((((.(((((......))).)))))).))))))).))..)))). ( -49.90) >DroAna_CAF1 266161 120 - 1 GGGCGACAAGAGGAAGUCCUUGCGGACGACGGUGCAGGGCUGGAUGCCCUGUUUGGUGGGCGCGUACUGCACCUUGCGGAUGCCCAGGUUGAUGGUGCUGCGGCGAUGCGACCGGUCGCA ..(((((..((((....)))).(((.(.((.(.(((((((.....))))))).).)).).(((((.(((((((....))..(((((......))).)))))))..))))).)))))))). ( -52.40) >consensus GGGCGACAAAAGGAAGUCCUUGCGCACCACCGUGCAGGGCUGCAUGCCCUGCUUGGUGGGUGCAUACUGCACCUUGCGGAUGCCCAGAUUGAUGGUGCUGCGACGAUGCGAACGAUCGCA (((((....((((....)))).((((.(((((.(((((((.....))))))).)))))(((((.....))))).))))..))))).....................(((((....))))) (-41.62 = -42.43 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:47 2006