| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,039,322 – 18,039,432 |
| Length | 110 |
| Max. P | 0.850236 |

| Location | 18,039,322 – 18,039,432 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
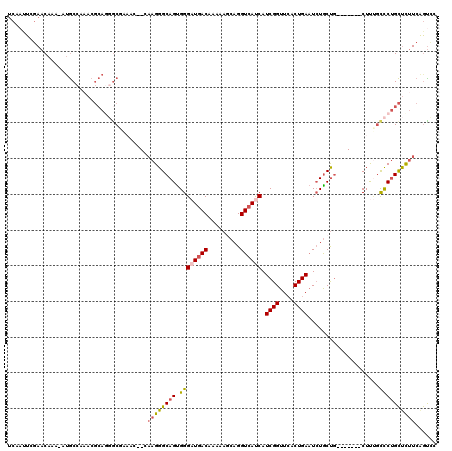
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.57 |
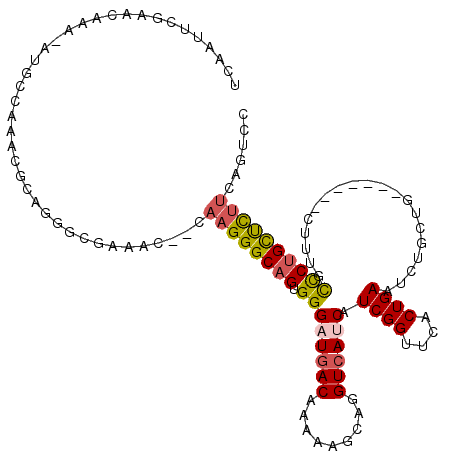
| Structure conservation index | 0.53 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18039322 110 - 22407834 UCAAUUCGAACAAA-AUGCCAAACGCAGGGCGAAAC--GAAGGGCAGUGGGAUGACAAAAAACAGGUCAUCAUCGGUUCACUGAAUCUACUG-------CUUUGCCCUGCUCUUCAGUCC ..............-.........((((((((((.(--....)(((((((((((((.........)))))).((((....))))..))))))-------))))))))))).......... ( -39.40) >DroSec_CAF1 255578 110 - 1 UCAAUUCGAACAAA-AUGCCAAACGCAGUGCGAAAC--GCAGGGCAGUGGGAUGACAAAAAGCAGGUCAUCAUCGGUUCACUGAAUAUGCUG-------CUUUGCCCUGCUCUUCAGUCC ....((((..((..-.(((.....))).))))))..--(((((((((..(((((((.........)))))).((((....)))).....)..-------)..)))))))).......... ( -33.40) >DroSim_CAF1 255608 110 - 1 UCAAUUCGAACAAA-AAGCCAAACGCAGGGCGAAAC--GAAGGGCAGUGGGAUGACAAAAAGCAGGUCAUCAUCGGUUCACUGAAUCUGCUG-------CUUUGCCCUGCUCUUCAGUCC ..............-.........((((((((((.(--....)((((..(((((((.........)))))).((((....))))..)..)))-------))))))))))).......... ( -38.50) >DroEre_CAF1 248628 105 - 1 UCAACUCGAACAAA-AUGCCAAACGCAGGGC-------CAAGGGCAGUGGGUUGACAAAAAGCAGGUCAUCAUCGGUUCACUGAAUCUGCUG-------CUCCGCCCUGCUUUUCGGCUG ..............-..((((((.(((((((-------...((((((..(((((((.........))))...((((....)))))))..)))-------))).))))))).))).))).. ( -40.40) >DroYak_CAF1 264830 104 - 1 UCAAUUUGAACAAA-AUGCCAAACGCAUGAC-------CAAGGGCAGUGGGAUGACAAAAAACAGGUCAUCAUCGGUUCACUGAAUCUGCUG-------CUUUGCCCUGCUC-UCGGUCC ..............-((((.....))))(((-------(.(((((((.((((((((.........)))))).((((....))))........-------.....))))))))-).)))). ( -32.70) >DroAna_CAF1 242168 118 - 1 GCAUCUCAAACAAAAAUACCAUAACGA--GCCCAACGGCAAGGGCAGCGGGAUCACAAAAAGCAGGUCACCAUCGGUUGACUGACUCUGCCCGCCCAAACUCGAUUCCGUCCUUCAAUCC ........................(((--(......(((..((((((.((((((..........)))).)).((((....))))..)))))))))....))))................. ( -26.60) >consensus UCAAUUCGAACAAA_AUGCCAAACGCAGGGCGAAAC__CAAGGGCAGUGGGAUGACAAAAAGCAGGUCAUCAUCGGUUCACUGAAUCUGCUG_______CUUUGCCCUGCUCUUCAGUCC .......................................((((((((.((((((((.........)))))).((((....))))....................))))))))))...... (-18.56 = -18.87 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:33 2006