| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,038,395 – 18,038,505 |
| Length | 110 |
| Max. P | 0.926963 |

| Location | 18,038,395 – 18,038,505 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.71 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -11.53 |
| Energy contribution | -12.62 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18038395 110 - 22407834 AAAUGGUAAAUGGUAAAUGGCGAAUGGCGAAUGGCGAAUGGUAAAU-GGCA----GCGGAUGGCAGCGCGUUGCUCAUACGCCAUGUG-GUGCCUGC-GGUGUGCCAAUAAUGCUGG ....((((..(((((.((.(((...((((.((((((.(((((((.(-(((.----((.....)).)).)))))).))).))))))...-.)))))))-.)).)))))....)))).. ( -36.40) >DroSec_CAF1 254668 86 - 1 AAAUGGUA---------------------AAUGGCGAAUGGUAAAU-GG-------CGGAUGGCAGCGCGUUGCUCAUACGCCAUGUG-GUGCCUGC-GGUGUGCCAAUAAUGCUGG ...(((((---------------------.((.(((...((((.((-((-------((.(((((((....)))).))).))))))...-.)))))))-.)).))))).......... ( -31.50) >DroSim_CAF1 254689 93 - 1 AAAUGGUAUA--------------UGGCGAAUGGCGAAUGGUAAAU-GG-------CGGAUGGCAGCGCGUUGCUCAUACGCCAUGUG-GUGCCUGC-GGUGUGCCAAUAAUGCUGG ...(((((((--------------(.(((...((((........((-((-------((.(((((((....)))).))).))))))...-.)))))))-.)))))))).......... ( -35.50) >DroEre_CAF1 247582 114 - 1 AAAUGGCAAAUGGUAAAUGGCGAAUGGUAAAUGGCGAAUGGUAAAU-GGCGGAUGGCGGAUGGCAGCGCGUUGCUCAUACGCCAUGUG-GUGCCUGC-GGUGUGCCAAUAAUGCUGG ....((((..(((((.((.(((...((((.((((((.(((((((..-.(((..((.(....).)).))).)))).))).))))))...-.)))))))-.)).)))))....)))).. ( -38.50) >DroWil_CAF1 322857 92 - 1 UACUUAUAUA--------------UGUUGUC-UUCCUUUGGAAAUCAUA--------ACAGCUUGAAAUGUUGCGCAUACGACAUGUGUGCGCCUCUUCCUACUCCAAUAAUCCA-- ..........--------------.......-.....(((((......(--------(((........))))((((((((.....))))))))..........))))).......-- ( -18.10) >DroYak_CAF1 264056 86 - 1 AAAUGGCGAA---------------------UAGCGAAUGGUAAAU-GG-------CGGAUGGCAGCGCGUUGCUCAUACGCCAUGUG-GUGCCUGC-GGUGUGCCAAUAAUGCUGG ...((((..(---------------------((.((...((((.((-((-------((.(((((((....)))).))).))))))...-.))))..)-).))))))).......... ( -31.60) >consensus AAAUGGUAAA______________UGG_GAAUGGCGAAUGGUAAAU_GG_______CGGAUGGCAGCGCGUUGCUCAUACGCCAUGUG_GUGCCUGC_GGUGUGCCAAUAAUGCUGG ...(((((......................((((((.(((.....................(((((....)))))))).))))))......(((....))).))))).......... (-11.53 = -12.62 + 1.09)

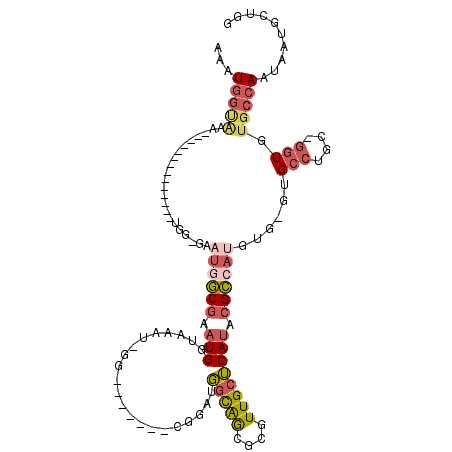

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:32 2006