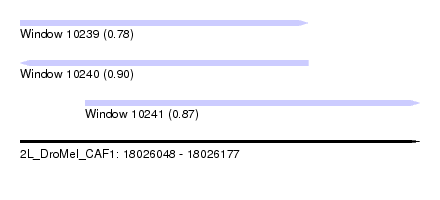
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,026,048 – 18,026,177 |
| Length | 129 |
| Max. P | 0.896503 |

| Location | 18,026,048 – 18,026,141 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -17.97 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18026048 93 + 22407834 AAAGCGAA-GGGAACAGCGGAAAUGAUGCGGCUCAUUUGUGUGCUACCACAAAAUGGUGGUGGCACAGAGGAAGUGGAAUAAAUGCAGUGAUUA ...(((..-(((....(((.......)))..))).(((.(((((((((((......))))))))))).)))............)))........ ( -28.00) >DroSec_CAF1 241963 93 + 1 AAAGCGAA-GGGAACAGCGGAAAUGAUGCGGCUCAUUUGUGUGCCACCACAAAAUGGUGGUGGCACAGGGGAAGUGGGAUAAGUGCAGUGGUUA ...((...-(....).))........((((.(((((((.(((((((((((......)))))))))))....))))))).....))))....... ( -30.80) >DroSim_CAF1 241972 93 + 1 AAAGCGAA-GGGAACAGCGGAAAUGAUGCGGCUCAUUUGUGUGCCACCACAAAAUGGUGGUGGCACAGAGGAAGUGGGAUAAGUGCAGUGAUUA ...((...-(....).))........((((.(((((((.(((((((((((......)))))))))))....))))))).....))))....... ( -30.80) >DroEre_CAF1 234567 94 + 1 AAAGCGAAGGGGAGAAGCGGAAGUGAUGCGGCUCAUUUGUGUGCGACCACAAAAUGGUGGUGGCACUGAGGAUGUGGCACAAAUGCAGUGAUUA ...(((....(.(...((....))..).).((((((((..((((.(((((......))))).))))...))))).))).....)))........ ( -29.20) >DroYak_CAF1 251345 94 + 1 AAAGCCAGAGGGAGAAGCGGAAAUGAUGCGGCUCAUUUGUGUGCGACCACAACAUGGUGGUGGCACUGAGGAUGUGGCACAAAUGCAGUGAUUA ...((((....(((..(((.......)))..)))((((..((((.(((((......))))).))))...)))).))))................ ( -29.80) >DroAna_CAF1 228847 84 + 1 AUAGAGAG-GGGGUGGCCGGAAGUGAUGCAGCUCAUUUGUGUCAGACCACAAAAUGGAGGAGAGAC--------UGCCAUA-AUGCAGGAAUUA ........-((..((((.(.((((((......)))))).)))))..)).................(--------(((....-..))))...... ( -16.80) >consensus AAAGCGAA_GGGAACAGCGGAAAUGAUGCGGCUCAUUUGUGUGCGACCACAAAAUGGUGGUGGCACAGAGGAAGUGGCAUAAAUGCAGUGAUUA ...(((((.(((....(((.......)))..))).)))))((((.(((((......))))).))))............................ (-17.97 = -18.47 + 0.50)



| Location | 18,026,048 – 18,026,141 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -14.63 |
| Energy contribution | -15.35 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18026048 93 - 22407834 UAAUCACUGCAUUUAUUCCACUUCCUCUGUGCCACCACCAUUUUGUGGUAGCACACAAAUGAGCCGCAUCAUUUCCGCUGUUCCC-UUCGCUUU ........((.................(((((.(((((......))))).))))).....((((.((.........)).))))..-...))... ( -20.40) >DroSec_CAF1 241963 93 - 1 UAACCACUGCACUUAUCCCACUUCCCCUGUGCCACCACCAUUUUGUGGUGGCACACAAAUGAGCCGCAUCAUUUCCGCUGUUCCC-UUCGCUUU ........((.................(((((((((((......))))))))))).....((((.((.........)).))))..-...))... ( -26.90) >DroSim_CAF1 241972 93 - 1 UAAUCACUGCACUUAUCCCACUUCCUCUGUGCCACCACCAUUUUGUGGUGGCACACAAAUGAGCCGCAUCAUUUCCGCUGUUCCC-UUCGCUUU ........((.................(((((((((((......))))))))))).....((((.((.........)).))))..-...))... ( -26.90) >DroEre_CAF1 234567 94 - 1 UAAUCACUGCAUUUGUGCCACAUCCUCAGUGCCACCACCAUUUUGUGGUCGCACACAAAUGAGCCGCAUCACUUCCGCUUCUCCCCUUCGCUUU ........((((((((............((((.(((((......))))).))))))))))(((..((.........))..)))......))... ( -18.50) >DroYak_CAF1 251345 94 - 1 UAAUCACUGCAUUUGUGCCACAUCCUCAGUGCCACCACCAUGUUGUGGUCGCACACAAAUGAGCCGCAUCAUUUCCGCUUCUCCCUCUGGCUUU ................((((........((((.(((((......))))).))))..((((((......)))))).............))))... ( -22.20) >DroAna_CAF1 228847 84 - 1 UAAUUCCUGCAU-UAUGGCA--------GUCUCUCCUCCAUUUUGUGGUCUGACACAAAUGAGCUGCAUCACUUCCGGCCACCCC-CUCUCUAU ......(((...-....(((--------(.(((........((((((......)))))).)))))))........))).......-........ ( -14.06) >consensus UAAUCACUGCAUUUAUGCCACUUCCUCUGUGCCACCACCAUUUUGUGGUCGCACACAAAUGAGCCGCAUCAUUUCCGCUGUUCCC_UUCGCUUU ........((..................((((.(((((......))))).))))..((((((......))))))..))................ (-14.63 = -15.35 + 0.72)

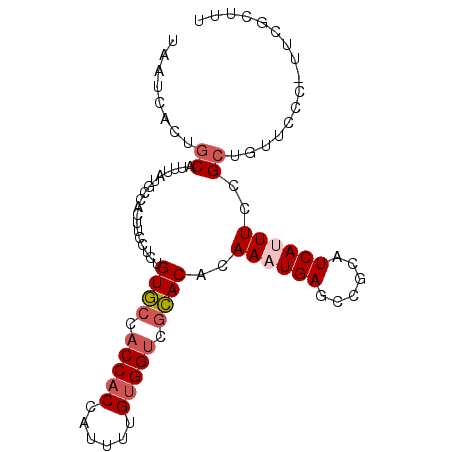

| Location | 18,026,069 – 18,026,177 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -21.52 |
| Energy contribution | -23.13 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 18026069 108 + 22407834 AUGAUGCGGCUCAUUUGUGUGCUACCACAAAAUGGUGGUGGCACAGAGGAAGUGGAAUAAAUGCAGUGAUUAAGGGGAGGACAGUUGGUUAACGCUUCCAAUUAGAAA ......(.(((..(((.(((((((((((......))))))))))).))).))).)..............((((..(((((...(((....))).)))))..))))... ( -32.70) >DroSec_CAF1 241984 108 + 1 AUGAUGCGGCUCAUUUGUGUGCCACCACAAAAUGGUGGUGGCACAGGGGAAGUGGGAUAAGUGCAGUGGUUAAGGGGAGGACAGUUGGUUAACGCUUCCAAUUAAAAA ....((((.(((((((.(((((((((((......)))))))))))....))))))).....))))....((((..(((((...(((....))).)))))..))))... ( -38.20) >DroSim_CAF1 241993 108 + 1 AUGAUGCGGCUCAUUUGUGUGCCACCACAAAAUGGUGGUGGCACAGAGGAAGUGGGAUAAGUGCAGUGAUUAAGGGGAGGACAGUUGGUUAACGCUUCCAAUUAAAAA ....((((.(((((((.(((((((((((......)))))))))))....))))))).....))))....((((..(((((...(((....))).)))))..))))... ( -38.30) >DroEre_CAF1 234589 108 + 1 GUGAUGCGGCUCAUUUGUGUGCGACCACAAAAUGGUGGUGGCACUGAGGAUGUGGCACAAAUGCAGUGAUUAAGGGGAGGACAGUUGGUAAACGCUUUCCAUUAGAAA ....((((((((((((..((((.(((((......))))).))))...))))).))).....))))....((((.((((((...(((....))).)))))).))))... ( -34.00) >DroYak_CAF1 251367 108 + 1 AUGAUGCGGCUCAUUUGUGUGCGACCACAACAUGGUGGUGGCACUGAGGAUGUGGCACAAAUGCAGUGAUUAAGGGGAGGACAGUUGGUAAACGCUUCCAAUUAGAAA ....((((((((((((..((((.(((((......))))).))))...))))).))).....))))....((((..(((((...(((....))).)))))..))))... ( -34.90) >DroPer_CAF1 338430 90 + 1 GCGAUG---CUCAUUUGUGUGCGACCACAAAAUGGGGGCUAAAUGGAGGCUAUGG---------AGGCGGUCAGGGGAAGA------GUAAAUGCUGGCGAUUAACUU ((....---(((((((.((((....)))))))))))((((.......))))....---------..)).(((((.(.....------.....).)))))......... ( -19.00) >consensus AUGAUGCGGCUCAUUUGUGUGCGACCACAAAAUGGUGGUGGCACAGAGGAAGUGGCAUAAAUGCAGUGAUUAAGGGGAGGACAGUUGGUAAACGCUUCCAAUUAAAAA ....((((..((((((.(((((((((((......))))))))))).))...))))......))))....((((..(((((...(((....))).)))))..))))... (-21.52 = -23.13 + 1.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:30 2006