| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 18,004,467 – 18,004,567 |
| Length | 100 |
| Max. P | 0.666602 |

| Location | 18,004,467 – 18,004,567 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -33.60 |
| Energy contribution | -35.10 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
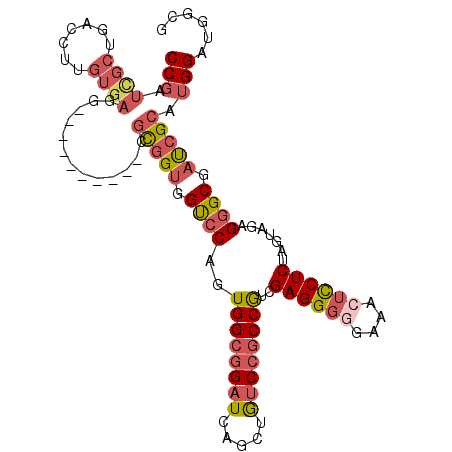
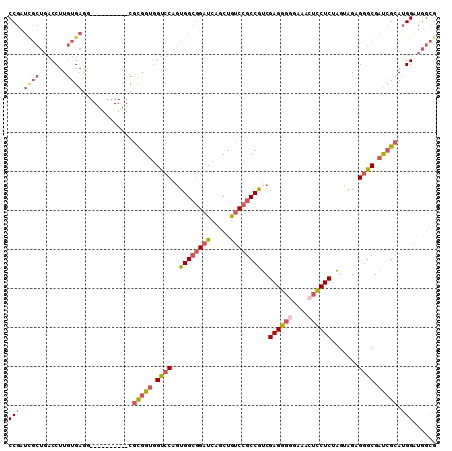
>2L_DroMel_CAF1 18004467 100 + 22407834 CCGAUCGCUGACCUUGUGAGG----------CGCGGUGGUCCAGUGGCGGAUCAGCUGUCCGCCGUCGAGGGCGAAACUCCUCUAGUAGAGGGCGAUCGCAUGGAUGGCG .....(((((.((.(((((..----------((((((.((((.(((((((((.....)))))))).)..))))...)))((((.....))))))).))))).)).))))) ( -43.10) >DroSec_CAF1 220443 100 + 1 CCGAUCGCUGACCUUGUGAGA----------CGCGGUGGUCCAGUGGCGGAUCAGCUGUCCGCCGUCGAGGGGGAAACUCCUCUAGUAGAGGGCGAUCGCAUGGAUGGCG .....(((((.((.(((((((----------((.(.((((((......)))))).)))))((((.((((((((....)))))).....)).)))).))))).)).))))) ( -47.40) >DroSim_CAF1 219513 100 + 1 CCGAUCGCUGACCUUGUGAGG----------CGCGGUGGUCCAGUGGCGGAUCAGCUAUCCGCCGUCGAGGGGGAAACUCCUCUAGUAGAGGGCGAUCGCAUGGAUGGCG (((.((((((((((((((...----------))))).))).)))))))))....((((((((((.((((((((....)))))).....)).)))(....)..))))))). ( -46.90) >DroEre_CAF1 211040 100 + 1 CCGAUUGCUGACCUUGUGACC----------CGCGGUGGUCCAGUGGCGGAUCAGCUGUCCGCCGUCGAGGGGGAAACUCCUCUAGUGGAGGGCGAUCGCAUGGAUGGCG ......((((.((.(((((..----------.....((((((......))))))(((.(((((....((((((....))))))..))))).)))..))))).)).)))). ( -45.00) >DroYak_CAF1 227758 100 + 1 CCGAUUGCUGACCUUGUGAGC----------CGUGGUGGUCCAGUGGCGGAUCAGCUGUCCGCCGUCGAGGGGGAAACUCCUCUAGUGGAGGGCGACCGCAUGGAUGGCG .((...(((.((...)).)))----------..))((.(((((...((((.((.(((.(((((....((((((....))))))..))))).))))))))).))))).)). ( -48.30) >DroAna_CAF1 210555 101 + 1 CC---------CCUUCUGCAGGGUGCUCCACUACGGCAGCCCCAUGGGCGAUCAGCUGGCUGCCACCGAGGGCAUAACAUCUCUGGUGGAGAGCGAUGGAAUGGACAAUG .(---------(.(((..((..((.((((((((.(((((((.....(((.....))))))))))...((((........)))))))))))).))..))))).))...... ( -37.80) >consensus CCGAUCGCUGACCUUGUGAGG__________CGCGGUGGUCCAGUGGCGGAUCAGCUGUCCGCCGUCGAGGGGGAAACUCCUCUAGUAGAGGGCGAUCGCAUGGAUGGCG (((.((((.......)))).............(((((.((((..((((((((.....))))))))..((((((....)))))).......)))).))))).)))...... (-33.60 = -35.10 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:22 2006