| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,999,647 – 17,999,741 |
| Length | 94 |
| Max. P | 0.831856 |

| Location | 17,999,647 – 17,999,741 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -13.86 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
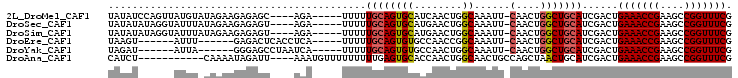
>2L_DroMel_CAF1 17999647 94 + 22407834 UAUAUCCAGUUAUGUAUAGAAGAGAGC----AGA-----UUUUUGCAGUGCAUCAACUGGCAAAUU-CAACUGGCUGCAUCGACUGAAACCGAAGCCGGUUUCG .....((((((((((((.(.(((((..----...-----))))).).)))))).))))))......-.((((((((...(((........)))))))))))... ( -25.60) >DroSec_CAF1 215720 94 + 1 UAUAUAUAGGUAUUUAUAGAAGAGAGU----AGA-----UUUUUGCAGUGCAUGAACUGGCAAAUU-CAACUGGCUGCAUCGACUGAAACCGAAGCCGGUUUCG ........(((.........((..(((----.((-----..((((((((......))).))))).)-).)))..))......)))(((((((....))))))). ( -21.66) >DroSim_CAF1 214293 94 + 1 UAUAUAUAGGUAUUUAUAGAAGAGAGU----AGA-----UUUUUGCAGUGCAUGAACUGGCAAAUU-CAACUGGCUGCAUCGACUGAAACCGAAGCCGGUUUCG ........(((.........((..(((----.((-----..((((((((......))).))))).)-).)))..))......)))(((((((....))))))). ( -21.66) >DroEre_CAF1 206323 86 + 1 UAAGU------AUUU------GAGACUCACCUCA-----UUUUUGCAGUGUGCCAACCGGCAAAUU-CAACUGGCUGCAUCGACUGAAACCGAAGCCGGUUUCG ..(((------...(------(((......))))-----....((((((.((((....))))....-......))))))...)))(((((((....))))))). ( -24.80) >DroYak_CAF1 222061 86 + 1 UAGAU------AUUA------GGGAGCCUAAUCA-----UUUUUGCAGUGUGCCAACUGGCAAAUU-CAACUGGCUGCAUCGACUGAAACCGAAGCCGGUUUCG ..(((------((((------((...))))))..-----.....(((((.((((....))))....-......))))))))....(((((((....))))))). ( -24.70) >DroAna_CAF1 205572 89 + 1 CAUCU-----------CAAAAUAGAUU----AAAUGUUUUUUUUUGAGUGCACCAACUGGCAACUGCCAGCUAACUGCAUCGACUGAAACCGAAGCCGGUUUCG ...((-----------(((((.(((..----......))).)))))))((((....(((((....))))).....))))......(((((((....))))))). ( -27.00) >consensus UAUAU______AUUU_UAGAAGAGAGU____AGA_____UUUUUGCAGUGCACCAACUGGCAAAUU_CAACUGGCUGCAUCGACUGAAACCGAAGCCGGUUUCG ...........................................((((((((........))......(....)))))))......(((((((....))))))). (-13.86 = -14.00 + 0.14)


| Location | 17,999,647 – 17,999,741 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.35 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
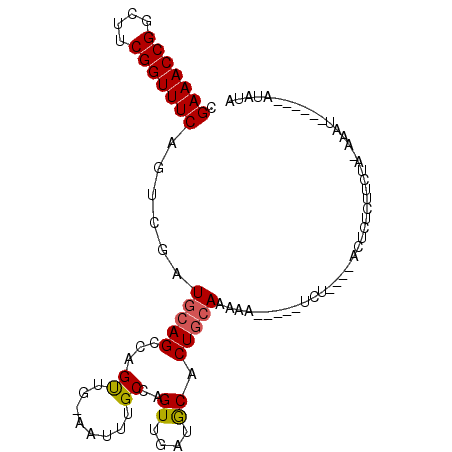
>2L_DroMel_CAF1 17999647 94 - 22407834 CGAAACCGGCUUCGGUUUCAGUCGAUGCAGCCAGUUG-AAUUUGCCAGUUGAUGCACUGCAAAAA-----UCU----GCUCUCUUCUAUACAUAACUGGAUAUA .(((((((....)))))))...........(((((((-....(((........)))..(((....-----..)----)).............)))))))..... ( -21.30) >DroSec_CAF1 215720 94 - 1 CGAAACCGGCUUCGGUUUCAGUCGAUGCAGCCAGUUG-AAUUUGCCAGUUCAUGCACUGCAAAAA-----UCU----ACUCUCUUCUAUAAAUACCUAUAUAUA .(((((((....)))))))(((.(((((((.....((-(((......)))))....)))))....-----)).----)))........................ ( -19.40) >DroSim_CAF1 214293 94 - 1 CGAAACCGGCUUCGGUUUCAGUCGAUGCAGCCAGUUG-AAUUUGCCAGUUCAUGCACUGCAAAAA-----UCU----ACUCUCUUCUAUAAAUACCUAUAUAUA .(((((((....)))))))(((.(((((((.....((-(((......)))))....)))))....-----)).----)))........................ ( -19.40) >DroEre_CAF1 206323 86 - 1 CGAAACCGGCUUCGGUUUCAGUCGAUGCAGCCAGUUG-AAUUUGCCGGUUGGCACACUGCAAAAA-----UGAGGUGAGUCUC------AAAU------ACUUA .(((((((....)))))))....((.((.(((((..(-.......)..))))).((((.((....-----)).)))).)).))------....------..... ( -24.40) >DroYak_CAF1 222061 86 - 1 CGAAACCGGCUUCGGUUUCAGUCGAUGCAGCCAGUUG-AAUUUGCCAGUUGGCACACUGCAAAAA-----UGAUUAGGCUCCC------UAAU------AUCUA .(((((((....)))))))......((((((((..((-.......))..))))....))))....-----..((((((...))------))))------..... ( -24.00) >DroAna_CAF1 205572 89 - 1 CGAAACCGGCUUCGGUUUCAGUCGAUGCAGUUAGCUGGCAGUUGCCAGUUGGUGCACUCAAAAAAAAACAUUU----AAUCUAUUUUG-----------AGAUG .(((((((....)))))))......((((.(((((((((....)))))))))))))(((((((..........----......)))))-----------))... ( -31.09) >consensus CGAAACCGGCUUCGGUUUCAGUCGAUGCAGCCAGUUG_AAUUUGCCAGUUGAUGCACUGCAAAAA_____UCU____ACUCUCUUCUA_AAAU______AUAUA .(((((((....)))))))......(((((...((........))..((....)).)))))........................................... (-13.38 = -13.35 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:21 2006