| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,999,448 – 17,999,547 |
| Length | 99 |
| Max. P | 0.846269 |

| Location | 17,999,448 – 17,999,547 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.43 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
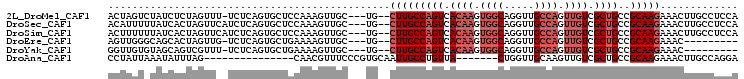
>2L_DroMel_CAF1 17999448 99 - 22407834 ACUAGUCUAUCUCUAGUUU-UCUCAGUGCUCCAAAGUUGC---UG--CUUGCCAGUCACAAGUGGCAGGUUGCCAGUUGUCGCUGCCGCAAGAAACUUGCCUCCA (((((.......)))))..-....((.((....(((((..---..--(((((((((.((((.((((.....)))).)))).))))..))))).)))))))))... ( -29.30) >DroSec_CAF1 215527 100 - 1 ACAUUUUUAUCACUAGUUCAUCUCAGUGCUCCAAAGUUGC---UG--CUUGCCAGUCACAAGUGGCAGGUUGCCAGUUGUCGCUGCCGCAAGAAACUUGCCUCCA ..........((((((.....)).)))).....(((((..---..--(((((((((.((((.((((.....)))).)))).))))..))))).)))))....... ( -28.50) >DroSim_CAF1 214093 100 - 1 ACUUUUUUAUCACUAGUUCAUCUCAGUGCUCCAAAGUUGC---UG--CUUGCCAGUCACAAGUGGCAGGUUGCCAGUUGUCGCUGCCGCAAGAAACUUGCCUCCA ..........((((((.....)).)))).....(((((..---..--(((((((((.((((.((((.....)))).)))).))))..))))).)))))....... ( -28.50) >DroEre_CAF1 206130 90 - 1 AGUUGGGCAGCACUAGUUG-UCUCAGUGCUGAAAAGUUGC---UG--CUUGCCAGUCACAAGUGGCAGGUUGCCAGUUGUCGCUGCCGCAAGAAAC--------- ....(((((((....))))-)))(((((((....)).)))---))--(((((((((.((((.((((.....)))).)))).))))..)))))....--------- ( -34.60) >DroYak_CAF1 221865 90 - 1 GGUUGUGUAGCAGUCGUUU-UCUCAGUGCUGAAAAGUUGC---UG--CUUGCCAGUCACAAGUGGCAGGUUGCCAGUUGUCGCUGCCGCAAGAAAC--------- (((...(((((((.(.(((-((........))))))))))---))--)..)))(((.((((.((((.....)))).)))).)))..(....)....--------- ( -31.10) >DroAna_CAF1 205400 83 - 1 CCUAUUAAAUAUUUAG---------------CAACGUUUCCCGUGCAAUUGCCUGUUA-------CUGGUUGCAAGUUGUCGCUGCCGCAAGAAACUUGCCAGGA ...............(---------------((.((.....))))).....((((...-------..(((.((........)).)))(((((...))))))))). ( -18.30) >consensus ACUUGUGUAUCACUAGUUC_UCUCAGUGCUCCAAAGUUGC___UG__CUUGCCAGUCACAAGUGGCAGGUUGCCAGUUGUCGCUGCCGCAAGAAACUUGCCUCCA ...............................................(((((((((.((((.((((.....)))).)))).))))..)))))............. (-17.10 = -18.43 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:19 2006