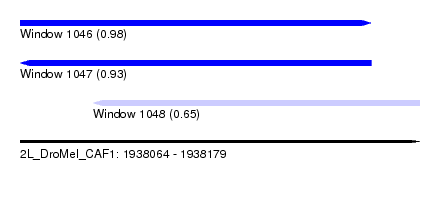
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,938,064 – 1,938,179 |
| Length | 115 |
| Max. P | 0.978301 |

| Location | 1,938,064 – 1,938,165 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -33.11 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1938064 101 + 22407834 ----------ACUAAAAA-CA---AAAAAAUUAAUUUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCGUG ----------........-..---...............((((((((((((.......)))))((((((.((...)).)))).)).................)))))))...... ( -32.20) >DroPse_CAF1 134994 108 + 1 ACAAUGAAA-----GAAA-CAAACAAAAAAUUAC-UUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUGUUG .........-----..((-(((..((((......-))))((((((((((((.......)))))((((((.((...)).)))).)).................)))))))))))). ( -32.90) >DroSim_CAF1 86698 100 + 1 ----------ACUAAAAA-CA---AAAAAGUUAU-UUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCGUG ----------........-..---..........-....((((((((((((.......)))))((((((.((...)).)))).)).................)))))))...... ( -32.20) >DroEre_CAF1 104915 102 + 1 -------AAAAUUAAAAACCA---AAAAAGUUCG-U--UGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCGUG -------..............---........((-.--.((((((((((((.......)))))((((((.((...)).)))).)).................)))))))..)).. ( -33.50) >DroWil_CAF1 146177 107 + 1 ------GAAAAACAGAAU-CAAAAAAAAAGAUAC-UUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUGUUG ------....(((((.((-(.........)))..-....((((((((((((.......)))))((((((.((...)).)))).)).................)))))))))))). ( -33.40) >DroPer_CAF1 133506 113 + 1 ACAAUGAAAAAGGAGAAA-CAAACAAAAAAUUAC-UUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUGUUG .(((..((((((......-..............)-))))((((((((((((.......)))))((((((.((...)).)))).)).................))))))))..))) ( -34.45) >consensus _______AA_ACUAAAAA_CA___AAAAAAUUAC_UUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCGUG .......................................((((((((((((.......)))))((((((.((...)).)))).)).................)))))))...... (-32.20 = -32.20 + -0.00)

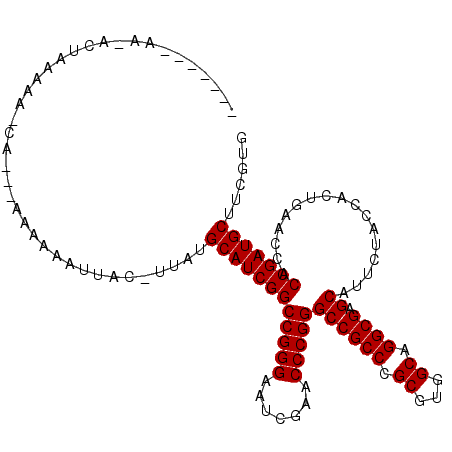
| Location | 1,938,064 – 1,938,165 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -36.93 |
| Consensus MFE | -35.90 |
| Energy contribution | -35.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1938064 101 - 22407834 CACGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAAAUUAAUUUUUU---UG-UUUUUAGU---------- ......(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...............---..-........---------- ( -35.90) >DroPse_CAF1 134994 108 - 1 CAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAA-GUAAUUUUUUGUUUG-UUUC-----UUUCAUUGU .((((((((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))((((-....)))))))))..-....-----......... ( -37.40) >DroSim_CAF1 86698 100 - 1 CACGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAA-AUAACUUUUU---UG-UUUUUAGU---------- ......(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).(((-(.(((.....---.)-))))))..---------- ( -36.40) >DroEre_CAF1 104915 102 - 1 CACGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCA--A-CGAACUUUUU---UGGUUUUUAAUUUU------- ..((..(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).--.-))((((....---.)))).........------- ( -37.90) >DroWil_CAF1 146177 107 - 1 CAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAA-GUAUCUUUUUUUUUG-AUUCUGUUUUUC------ .(((..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...(-(.(((.........)-)).)))))....------ ( -36.60) >DroPer_CAF1 133506 113 - 1 CAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAA-GUAAUUUUUUGUUUG-UUUCUCCUUUUUCAUUGU .((((((((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))((((-....)))))))))..-.................. ( -37.40) >consensus CAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAA_GUAACUUUUU___UG_UUUCUAGU_UU_______ ......(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))....................................... (-35.90 = -35.90 + -0.00)

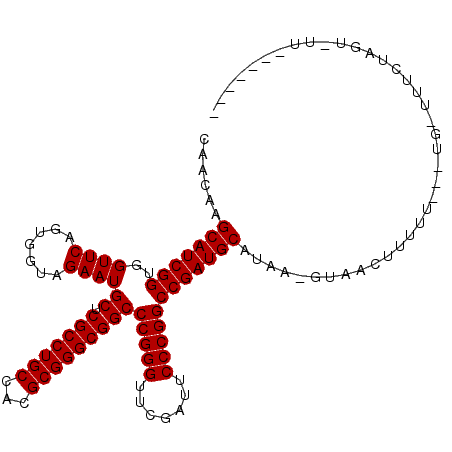
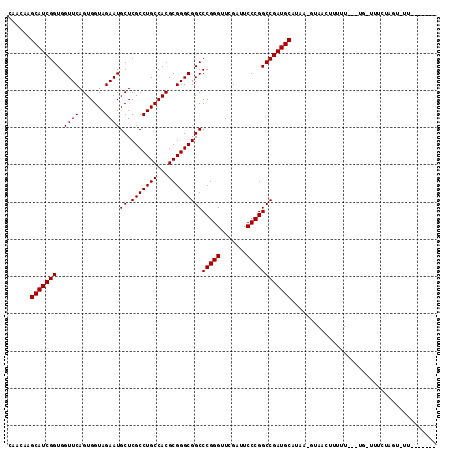
| Location | 1,938,085 – 1,938,179 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -35.20 |
| Energy contribution | -35.37 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1938085 94 - 22407834 ------------GAUU-UUC----------AUUG---ACUCACGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAA ------------....-.((----------((((---(..(((((....)).)))..)))))))....((((((((((((...))))))((((.(((.......))))))))).)))).. ( -38.20) >DroPse_CAF1 135022 109 - 1 CGAGAUUGGUGUUAUU-UUU----------GGGAAACUUUCAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAA .((((((((((((...-...----------((....)).......)))))))))..))).(((((((.........)))))))(((..(((((.(((.......)))))))).))).... ( -39.52) >DroEre_CAF1 104939 92 - 1 ------------GGUU-UUC----------CUGG---AGUCACGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCA--A ------------.(((-(((----------(..(---(..(((((....)).)))..))...)).)))))((((((((((...))))))((((.(((.......))))))))).)).--. ( -36.60) >DroWil_CAF1 146204 105 - 1 ------------GAGUGCUCCUAUAGGCAGAUCC---ACCCAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAA ------------...((((......))))((.((---(((...........))))).)).(((((((.........)))))))(((..(((((.(((.......)))))))).))).... ( -40.40) >DroAna_CAF1 109064 94 - 1 ------------GGAU-UGG----------ACUC---UUCCAAGAAGCCUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAA ------------(.((-(((----------..((---(.(((...((((.....))))...))).)))..((.(((((((...)))))))))(((((.......)))))))))).).... ( -38.40) >DroPer_CAF1 133539 109 - 1 CGAGAUUGGUGUUAUU-UUU----------GGGAAACUUUCAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAA .((((((((((((...-...----------((....)).......)))))))))..))).(((((((.........)))))))(((..(((((.(((.......)))))))).))).... ( -39.52) >consensus ____________GAUU_UUC__________AUGA___AUUCAACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAA ..............................................(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).... (-35.20 = -35.37 + 0.17)

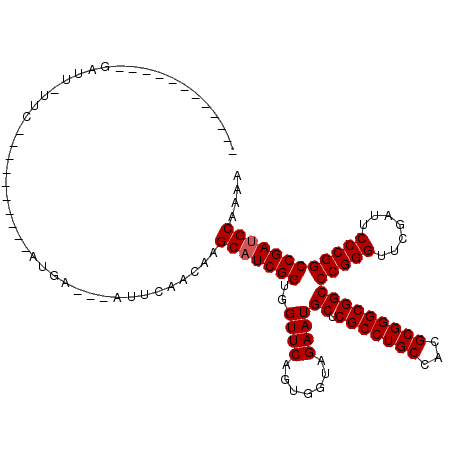
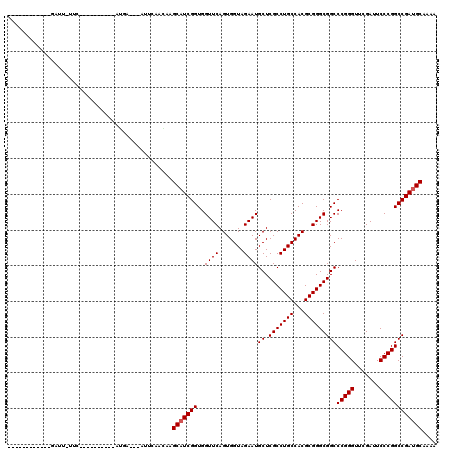
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:42 2006