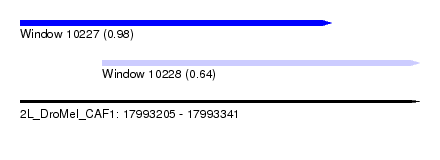
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,993,205 – 17,993,341 |
| Length | 136 |
| Max. P | 0.984905 |

| Location | 17,993,205 – 17,993,311 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.02 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -19.07 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
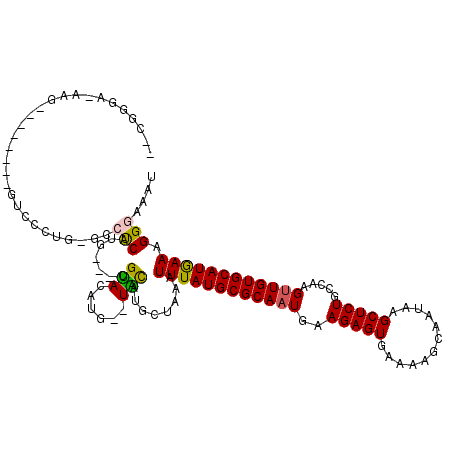
>2L_DroMel_CAF1 17993205 106 + 22407834 UACGGGAAAAG--------GUCCCUG-GCCUCGUG---CGUACAUG--UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAU ...........--------.((((((-((...(((---((....))--)))..)))...((((((((((((..(((((............))))).....)))))))))))))))))... ( -31.90) >DroVir_CAF1 269668 87 + 1 ---------------------------AUAUCAUG---CGUAU-UG--UAUAUGCUAAAUUUAUGCGCAAUGAAGAGUAAAAAGCAAUAAGCUCUGUUAAGUUGUGCAUAAAAGGGAAAU ---------------------------...((..(---(((((-..--.))))))....((((((((((((..(((((............))))).....))))))))))))...))... ( -22.30) >DroGri_CAF1 285548 92 + 1 ---------------------------ACAUCAUGCUGCGCAU-UGUGUGUAUGCUAAAUUUAUGCGCACUGAAGAGUGAAAAGCAAUAAGCUCUGUUAAGUUGUGCAUAAAAGGGAAAU ---------------------------...((..(((((((..-...))))).))....((((((((((((..(((((............)))))....)).))))))))))...))... ( -24.40) >DroEre_CAF1 199849 105 + 1 GCCGGGA-AAG--------GUCCCUG-GCCUCUUG---CGUACAUG--UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAU ((((((.-...--------...))))-))((((((---(((.....--...)))).....(((((((((((..(((((............))))).....)))))))))))))))).... ( -33.90) >DroYak_CAF1 215127 105 + 1 GUCGUGA-AAG--------GUCCCUG-GCCUCUUG---CGUACAUG--UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAU ((((..(-.((--------((.....-)))).)..---)).))...--......((...((((((((((((..(((((............))))).....))))))))))))..)).... ( -30.30) >DroAna_CAF1 199376 114 + 1 UCUGGGAGGGGAGCGACUUAUCGCUGGGACCCCUG---GGUAUG-G--UGUAUGCUGAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAU (((...((((((((((....)))))....))))).---((((((-.--..))))))...((((((((((((..(((((............))))).....))))))))))))..)))... ( -34.80) >consensus __CGGGA_AAG________GUCCCUG_GCCUCAUG___CGUACAUG__UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAU .............................(((.......(((......)))........((((((((((((..(((((............))))).....)))))))))))).))).... (-19.07 = -18.60 + -0.47)


| Location | 17,993,233 – 17,993,341 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -20.63 |
| Energy contribution | -20.91 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
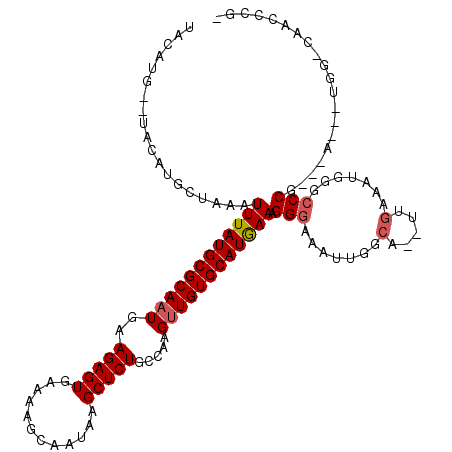
>2L_DroMel_CAF1 17993233 108 + 22407834 UACAUG--UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAUUGGCA--UUGAAAUGGGCCCG----U--UGG-CAACCCG- ......--....((((((.((((((((((((..(((((............))))).....)))))))))))).(((..(((....--....)))...))).----)--)))-)).....- ( -30.50) >DroVir_CAF1 269678 99 + 1 UAU-UG--UAUAUGCUAAAUUUAUGCGCAAUGAAGAGUAAAAAGCAAUAAGCUCUGUUAAGUUGUGCAUAAAAGGGAAAUUGGCA--UUGACAUGGGGCCAGGG---------------- ...-((--(..(((((((.((((((((((((..(((((............))))).....)))))))))))).......))))))--)..)))...........---------------- ( -23.80) >DroGri_CAF1 285561 116 + 1 CAU-UGUGUGUAUGCUAAAUUUAUGCGCACUGAAGAGUGAAAAGCAAUAAGCUCUGUUAAGUUGUGCAUAAAAGGGAAAUAGGCAUAUUGACAUGGGCCCAUCAAC--UGA-CAACUAGC ((.-(((((.(((.((...((((((((((((..(((((............)))))....)).))))))))))..))..))).))))).))..(((....)))....--...-........ ( -26.40) >DroEre_CAF1 199876 108 + 1 UACAUG--UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAUUGGCA--UUAAAAUGGGCCCG---A---UGG-CAGCCCG- ......--....(((((..((((((((((((..(((((............))))).....)))))))))))).(((..(((....--....)))...))).---.---)))-)).....- ( -29.70) >DroYak_CAF1 215154 108 + 1 UACAUG--UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAUUGGCA--UUGAAAUGGGCCCG---A---UGG-CAACCCG- ......--...........((((((((((((..(((((............))))).....)))))))))))).(((...(((.((--(((.........))---)---)).-)))))).- ( -29.90) >DroAna_CAF1 199413 113 + 1 UAUG-G--UGUAUGCUGAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAUUGGCA--UUGACAUGGGCCCCCG-AAUAUGGAGGAAUCG- ....-(--(((((((..(.((((((((((((..(((((............))))).....)))))))))))).......)..)))--)..))))((..((((.-.....)).))..)).- ( -28.70) >consensus UACAUG__UACAUGCUAAAUUUAUGCGCAAUGAAGAGUGAAAAGCAAUAAGCUCUGCCAAGUUGUGCAUGAAAGGGAAAUUGGCA__UUGAAAUGGGCCCG___A___UGG_CAACCCG_ ...................((((((((((((..(((((............))))).....)))))))))))).(((.......(.....).......))).................... (-20.63 = -20.91 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:18 2006