
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,990,931 – 17,991,038 |
| Length | 107 |
| Max. P | 0.828102 |

| Location | 17,990,931 – 17,991,038 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.64 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
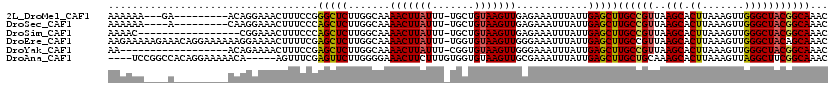
>2L_DroMel_CAF1 17990931 107 + 22407834 AAAAAA---GA---------ACAGGAAACUUUCCGGGCUCUUGGCAAAACUUAUUU-UGCUGUAAGUUGAGAAAUUUAUUGAGCUUGCCGUUAAGCACUUAAAGUUGGGCUACGGCAAAC ......---((---------(.((....))))).((((((..(((((((....)))-))))(((((((....))))))).))))))(((((..(((.((.......)))))))))).... ( -29.40) >DroSec_CAF1 207131 106 + 1 AAAAAA----A---------CAAGGAAACUUUCCCAGCUCUUGGCAAAACUUAUUU-UGCUGUAAGUUGAGAAAUUUAUUGAGCUUGCCGUUAAGCACUUAAAGUUGGGCUACGGCAAAC ......----.---------((((....)((((.(((((..((((((((....)))-)))))..))))).))))....)))...(((((((..(((.((.......)))))))))))).. ( -29.40) >DroSim_CAF1 205695 102 + 1 AAAAC-----------------CGGAAACUUUCCCAGCUCUUGGCAAAACUUAUUU-UGCUGUAAGUUGAGAAAUUUAUUGAGCUUGCCGUUAAGCACUUAAAGUUGGGCUACGGCAAAC .....-----------------.(....)((((.(((((..((((((((....)))-)))))..))))).))))..........(((((((..(((.((.......)))))))))))).. ( -28.90) >DroEre_CAF1 197557 119 + 1 AAGAAAAAGAAACAGGAAAAAAGGAAAACUUUUCGAGCUCUUGGCAAAACUUAUUU-UGGUGUAAGUUGGGAAAUUUAUUGAGCUUGCCGUUAAGCACUUAAAGUUGGGCUACAGCAAAC ..........(((.((..(((((.....)))))(((((((....(..(((((((..-....)))))))..).........))))))))))))...........((((.....)))).... ( -21.52) >DroYak_CAF1 212862 101 + 1 AA------------------ACAGAAAACUUUCCGAGCUCUUGGCAAAACUUAUUU-CGGUGUAAGUUGGGAAAUUUAUUGAGCUUGCCGUUAAGCACUUAAAGUUGGGCUACGGCAAAC ..------------------.(((.(((.(((((..((.....))..(((((((..-....)))))))))))).))).)))...(((((((..(((.((.......)))))))))))).. ( -25.90) >DroAna_CAF1 197164 111 + 1 ----UCCGGCCACAGGAAAAACA-----AGUUUCGAGUUCUUGGGGAAACUUCUUUGUGGUGUAAGUUGCGAAAUUUAUUGAGCUUGCUGCAAAGCACUUAAAGUUAGGCUUCGGCAAAC ----....(((((((((....((-----((.........))))((....)))).)))))))(((((((.(((......))))))))))(((.((((.((.......))))))..)))... ( -27.30) >consensus AAAAAA____A_________ACAGGAAACUUUCCGAGCUCUUGGCAAAACUUAUUU_UGCUGUAAGUUGAGAAAUUUAUUGAGCUUGCCGUUAAGCACUUAAAGUUGGGCUACGGCAAAC ...................................(((((.......(((((((.......)))))))............)))))((((((..(((.((.......)))))))))))... (-18.67 = -18.64 + -0.02)
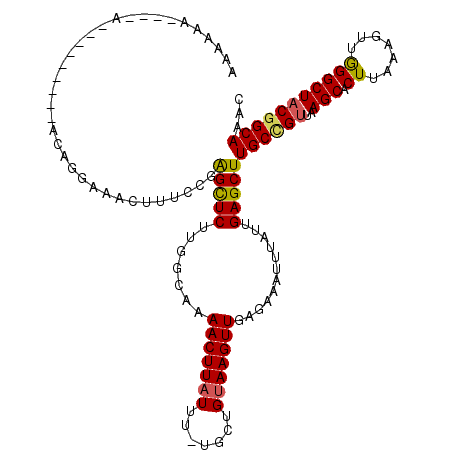

| Location | 17,990,931 – 17,991,038 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.828102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
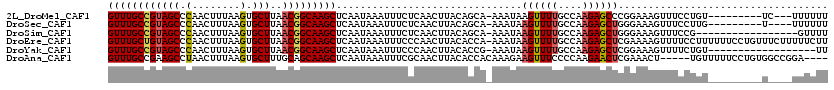
>2L_DroMel_CAF1 17990931 107 - 22407834 GUUUGCCGUAGCCCAACUUUAAGUGCUUAACGGCAAGCUCAAUAAAUUUCUCAACUUACAGCA-AAAUAAGUUUUGCCAAGAGCCCGGAAAGUUUCCUGU---------UC---UUUUUU ((((((((((((.(........).)))..)))))))))......................(((-(((....)))))).((((((..(((.....))).))---------))---)).... ( -26.50) >DroSec_CAF1 207131 106 - 1 GUUUGCCGUAGCCCAACUUUAAGUGCUUAACGGCAAGCUCAAUAAAUUUCUCAACUUACAGCA-AAAUAAGUUUUGCCAAGAGCUGGGAAAGUUUCCUUG---------U----UUUUUU ((((((((((((.(........).)))..)))))))))........(((((((.(((...(((-(((....))))))...))).))))))).........---------.----...... ( -29.00) >DroSim_CAF1 205695 102 - 1 GUUUGCCGUAGCCCAACUUUAAGUGCUUAACGGCAAGCUCAAUAAAUUUCUCAACUUACAGCA-AAAUAAGUUUUGCCAAGAGCUGGGAAAGUUUCCG-----------------GUUUU ((((((((((((.(........).)))..)))))))))........(((((((.(((...(((-(((....))))))...))).))))))).......-----------------..... ( -29.00) >DroEre_CAF1 197557 119 - 1 GUUUGCUGUAGCCCAACUUUAAGUGCUUAACGGCAAGCUCAAUAAAUUUCCCAACUUACACCA-AAAUAAGUUUUGCCAAGAGCUCGAAAAGUUUUCCUUUUUUCCUGUUUCUUUUUCUU ((((((((((((.(........).)))..)))))))))..............((((((.....-...)))))).....((((((..((((((.......))))))..).)))))...... ( -20.20) >DroYak_CAF1 212862 101 - 1 GUUUGCCGUAGCCCAACUUUAAGUGCUUAACGGCAAGCUCAAUAAAUUUCCCAACUUACACCG-AAAUAAGUUUUGCCAAGAGCUCGGAAAGUUUUCUGU------------------UU ((((((((((((.(........).)))..))))))))).((..((((((.((((((((.....-...))))))..((.....))..)).))))))..)).------------------.. ( -22.80) >DroAna_CAF1 197164 111 - 1 GUUUGCCGAAGCCUAACUUUAAGUGCUUUGCAGCAAGCUCAAUAAAUUUCGCAACUUACACCACAAAGAAGUUUCCCCAAGAACUCGAAACU-----UGUUUUUCCUGUGGCCGGA---- ((((((((((((((.......)).))))))..))))))......................(((((((.(((((((...........))))))-----).)).....))))).....---- ( -23.80) >consensus GUUUGCCGUAGCCCAACUUUAAGUGCUUAACGGCAAGCUCAAUAAAUUUCUCAACUUACACCA_AAAUAAGUUUUGCCAAGAGCUCGGAAAGUUUCCUGU_________U____UUUUUU ((((((((((((.(........).)))..)))))))))...............................((((((....))))))................................... (-15.72 = -16.00 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:16 2006