
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,988,101 – 17,988,202 |
| Length | 101 |
| Max. P | 0.845265 |

| Location | 17,988,101 – 17,988,202 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -15.08 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
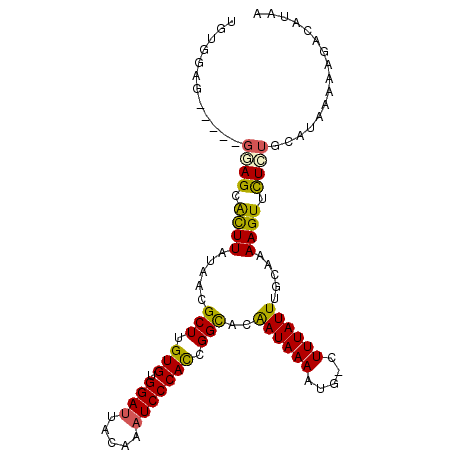
>2L_DroMel_CAF1 17988101 101 + 22407834 UGUGGAGG-AAGGGAGCGCUUAUAACGCUUGUGUGGAUUACAAAUCCCACCGGUGCGAUAAAAUG-GUUUAUUUGCAAAAGUUCUCUGCAUAAAAAUACAUAA ((..((((-(.(((((((.......)))(((((.....))))).)))).....(((((((((...-.)))).)))))....)))))..))............. ( -25.00) >DroVir_CAF1 262562 97 + 1 GAAGGAA---GAGCAGCAUUUAUAACGCUUGUGUGGAUUAUAAAUCCCACCGGCACAAUAAACUG-CUUUAUUUGCAAAAGUUUUGUGCCUGAAAGGGCAC-- ......(---((((((..(((((...(((.(((.((((.....))))))).)))...))))))))-))))...............((((((....))))))-- ( -30.30) >DroEre_CAF1 194748 97 + 1 UGUGGAG-----GGAGCACUUAUAACGCUUGUGUGGAUUACAAAUCCCACCGGUGCGAUAAAAUG-GUUUAUUUGCAAAAGUUCUCUGCAUAAAAAUACAUAA ((((.((-----(((((..((((..((((.(((.((((.....))))))).))))..))))....-((......))....))))))).))))........... ( -27.00) >DroYak_CAF1 210003 94 + 1 UGUGG---------AGCACUUAUAACGCUUGUGUGGAUUACAAAUCCCACCGGCGCGAUAAAAUGGGUUUAUUUGCAAAAGUUCUCUGCAUAAAAAUACGUAA ((..(---------((.((((.....(((.(((.((((.....))))))).)))((((((((.....)))).))))..)))).)))..))............. ( -26.00) >DroMoj_CAF1 309349 97 + 1 UGUAGAG---GCGCAGCAUUUAUAACGCUUGUGUGGAUUAUAAAUCCCACUGGCACAAUAAACUG-CUUUAUUUGCAGAAGUUUUGUGCAUAAAAAGGCAC-- ((((((.---((...)).))))))..(((((((.((((.....)))))))..((((((....(((-(.......)))).....))))))......))))..-- ( -26.20) >DroAna_CAF1 194624 102 + 1 GCAAAAGGUGUGGGAGAACUUAUAACGCUUGUGUGGAUUACCAUUCCCAUGGGCACAAUAAACUG-CUUUAUUUGCGAAAGUUCUGUGCAUGAAAGGACUACA (((((.(.((((((....)))))).)(((..((.(((.......)))))..)))...........-.....)))))...((((((.........))))))... ( -24.50) >consensus UGUGGAG_____GGAGCACUUAUAACGCUUGUGUGGAUUACAAAUCCCACCGGCACAAUAAAAUG_CUUUAUUUGCAAAAGUUCUCUGCAUAAAAAGACAUAA ............((((.((((.....(((.(((.((((.....))))))).)))..((((((.....)))))).....)))).))))................ (-15.08 = -14.33 + -0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:12 2006