| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,987,853 – 17,987,944 |
| Length | 91 |
| Max. P | 0.879757 |

| Location | 17,987,853 – 17,987,944 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -9.51 |
| Energy contribution | -9.84 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17987853 91 + 22407834 CGGCCAACAGCAGCAAUAAUAAACAAGAAAGUGUGAAAUAAGGCCAAAUGGACAUUC-AAAUUUGUAUGUCCAACACGAG----AAA----CCG-GCAGCA ..(((.......((........(((......)))........))....(((((((.(-(....)).))))))).......----...----..)-)).... ( -17.39) >DroVir_CAF1 262259 98 + 1 -GGUCGAGCACUGCAAUAAUAAACAAGAAAGUGUGAAAUAAAGCGAAAUGGACAUAACAAAUUUGUAUGUCCAGCGACAGUGGGAAA-GA-GUGAAAAAUA -.((((.(((((.................)))))..............((((((((.((....)))))))))).)))).........-..-.......... ( -19.43) >DroPse_CAF1 291674 87 + 1 CGG--AGCCCCAGCAAUAAUAAACAAGAAAGUGUGAAAUAAAGCCAAAUGGACAUAA-AAAUUUGUAUGUCCAACAAGAG----GAG------G-GCGGCA .(.--.(((((.((........(((......)))........))....((((((((.-.......)))))))).......----).)------)-))..). ( -19.39) >DroEre_CAF1 194492 91 + 1 CGGCCAACAGCAGCAAUAAUAAACAAGAAAGUGUGAAAUAAGGCCAAAUGGACAUAC-AAAUUUGUAUGUCCAACACGAG----AAA----CUG-GCAGCA ..((((......((........(((......)))........))....(((((((((-(....)))))))))).......----...----.))-)).... ( -22.49) >DroAna_CAF1 194337 95 + 1 AGGAGCACAACGGCAAUAAUAAACAAGAAAGUGUGAAAUAAAGCCAAAUGGACAUAA-AAAUUUGUAUGUCCUACAAGAG----GAAGUAGUCG-GCUGCG .(.(((.(.(((((........(((......)))........)))....(((((((.-.......)))))))........----......)).)-))).). ( -19.89) >DroPer_CAF1 293013 87 + 1 CGG--AGCCCCAGCAAUAAUAAACAAGAAAGUGUGAAAUAAAGCCAAAUGGACAUAA-AAAUUUGUAUGUCCAACAAGAG----GAG------G-GCGGCA .(.--.(((((.((........(((......)))........))....((((((((.-.......)))))))).......----).)------)-))..). ( -19.39) >consensus CGG_CAACAACAGCAAUAAUAAACAAGAAAGUGUGAAAUAAAGCCAAAUGGACAUAA_AAAUUUGUAUGUCCAACAAGAG____AAA______G_GCAGCA ............((........(((......)))........))....((((((((.........))))))))............................ ( -9.51 = -9.84 + 0.33)

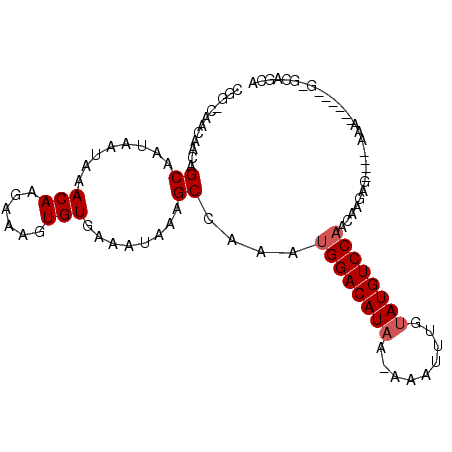

| Location | 17,987,853 – 17,987,944 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -10.64 |
| Energy contribution | -10.78 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17987853 91 - 22407834 UGCUGC-CGG----UUU----CUCGUGUUGGACAUACAAAUUU-GAAUGUCCAUUUGGCCUUAUUUCACACUUUCUUGUUUAUUAUUGCUGCUGUUGGCCG .(((..-(((----(..----.......(((((((.((....)-).)))))))...(((..((....(((......)))....))..)))))))..))).. ( -18.60) >DroVir_CAF1 262259 98 - 1 UAUUUUUCAC-UC-UUUCCCACUGUCGCUGGACAUACAAAUUUGUUAUGUCCAUUUCGCUUUAUUUCACACUUUCUUGUUUAUUAUUGCAGUGCUCGACC- ..........-..-.....((((((.((((((((((((....)).))))))))....))........(((......)))........)))))).......- ( -16.40) >DroPse_CAF1 291674 87 - 1 UGCCGC-C------CUC----CUCUUGUUGGACAUACAAAUUU-UUAUGUCCAUUUGGCUUUAUUUCACACUUUCUUGUUUAUUAUUGCUGGGGCU--CCG ....((-(------((.----.......((((((((.......-.))))))))...(((..((....(((......)))....))..)))))))).--... ( -21.40) >DroEre_CAF1 194492 91 - 1 UGCUGC-CAG----UUU----CUCGUGUUGGACAUACAAAUUU-GUAUGUCCAUUUGGCCUUAUUUCACACUUUCUUGUUUAUUAUUGCUGCUGUUGGCCG .(((..-(((----(..----.......((((((((((....)-)))))))))...(((..((....(((......)))....))..)))))))..))).. ( -23.90) >DroAna_CAF1 194337 95 - 1 CGCAGC-CGACUACUUC----CUCUUGUAGGACAUACAAAUUU-UUAUGUCCAUUUGGCUUUAUUUCACACUUUCUUGUUUAUUAUUGCCGUUGUGCUCCU .(.(((-((((......----........(((((((.......-.)))))))....(((..((....(((......)))....))..))))))).))).). ( -21.80) >DroPer_CAF1 293013 87 - 1 UGCCGC-C------CUC----CUCUUGUUGGACAUACAAAUUU-UUAUGUCCAUUUGGCUUUAUUUCACACUUUCUUGUUUAUUAUUGCUGGGGCU--CCG ....((-(------((.----.......((((((((.......-.))))))))...(((..((....(((......)))....))..)))))))).--... ( -21.40) >consensus UGCUGC_C______UUC____CUCUUGUUGGACAUACAAAUUU_UUAUGUCCAUUUGGCUUUAUUUCACACUUUCUUGUUUAUUAUUGCUGCGGUUG_CCG .(((.........................(((((((.........)))))))....(((..((....(((......)))....))..)))..)))...... (-10.64 = -10.78 + 0.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:11 2006