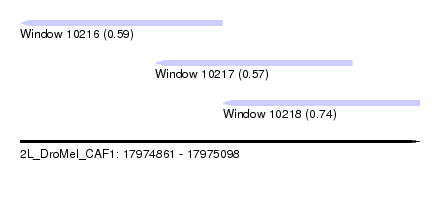
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,974,861 – 17,975,098 |
| Length | 237 |
| Max. P | 0.744708 |

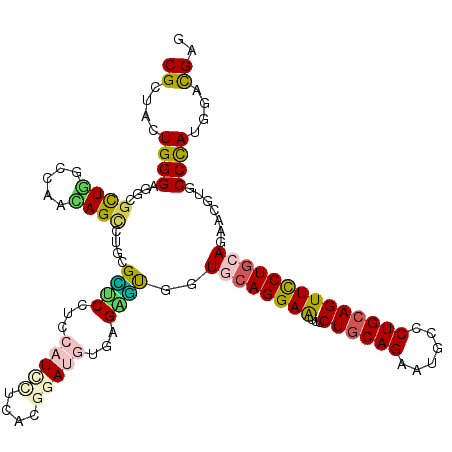
| Location | 17,974,861 – 17,974,981 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -41.87 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.17 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17974861 120 - 22407834 GGUAGCAUUAGUCAAUCGGGAAUGAAGUACGAGAAGAUAAUGAGUAAAUUGCCGCAGAUCGAGAAAACGUUGGAGGCGGUGCGAGCAAAGGAUUCCUGUCUGGGGGGCUGGUGCAAGGAA ....(((((((((...(((((((....(((.............)))....(((((...((.((......)).)).)))))...........))))))).......)))))))))...... ( -30.02) >DroVir_CAF1 247566 120 - 1 GGCAGCAUCAGUCAGUCGGGCAUGAAGUAUGAGAAGAUCAUGAGCGAGCUGCCGCAGAUCAGCAGCACACUGGGGCUGGUGCGUGAGAAGGAUGCCUGCCUGGCGGGCUGGUGCAAGGAG ....(((((((((.(((((((......(((((.....)))))((.(.(((((.........))))).).)).((((.....(....)......))))))))))).)))))))))...... ( -47.60) >DroGri_CAF1 260672 120 - 1 GGCAGCAUCAGUGAGUCGGGCAUGAAGUAUGAGAAUAUUAUGCGCGAGUUGCCACAGAUCAGUGGCACGUUGGAGUUGGUCCGGCAGAAGGAUGCCUGCUUGGCGGGGUGGUGCAAGGAA .(((.((((.((.((.(((((((...((((((.....))))))....((.(((((......)))))))((((((.....))))))......))))))).)).))..)))).)))...... ( -44.70) >DroMoj_CAF1 293595 120 - 1 GGCAGCAUCAGUGAGUCGGGCAUGAAGUACGAGAAGAUCAUGAGCGAGCUGCCACAGAUCAGCAGCACGCUGGAGCUGGUCAGGGCCAAGGAUGCCUGCCUGGCGGGCUGGUGCAAGGAG ....((((((((..(((((((..............((((((.((((.(((((.........))))).)))).)...))))).((((.......)))))))))))..))))))))...... ( -50.70) >DroAna_CAF1 181534 120 - 1 GGCAGCAUCAGCCAGUCGGGAAUGAAGUACGAAAAGAUAAUGACCGAGCUGCCGCAGAUCGCCAAGACCCUGGACGCUGUGCGAGAGAAGGACGCUUGCCUGGCAGGAUGGUGCAAGGAA .(((.((((......((((..((................))..)))).(((((((((....(((......)))...))))(((((.(.....).)))))..))))))))).)))...... ( -37.19) >DroPer_CAF1 281976 120 - 1 GGUAGCAUCAGUCAGUCAGGCAUGAAGUACGAGAAGAUCAUGAGCGAGCUGCCGCAGAUCGGAGAGACACUUGAAGUGGUGAGGGCCAAAGAUGCCUGUCUGGCGGGCUGGUGCAAGGAG ....(((((((((.(((((((..............((((....(((......))).)))).......((((......)))).((((.......))))))))))).)))))))))...... ( -41.00) >consensus GGCAGCAUCAGUCAGUCGGGCAUGAAGUACGAGAAGAUCAUGAGCGAGCUGCCGCAGAUCAGCAACACGCUGGAGGUGGUGCGGGAGAAGGAUGCCUGCCUGGCGGGCUGGUGCAAGGAA ....(((((((((.((((((((...(((.((...(.....)...)).)))((((....((((.......))))...))))................)))))))).)))))))))...... (-25.74 = -25.17 + -0.58)

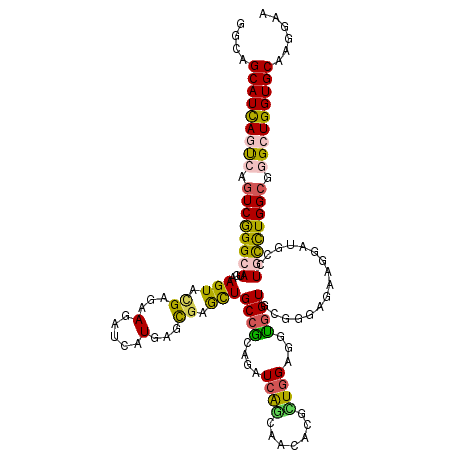

| Location | 17,974,941 – 17,975,058 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.99 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -29.28 |
| Energy contribution | -28.27 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17974941 117 - 22407834 UUUUGACGGAUGUGAGUUUAAUCCAGGAAUUCCUACAGAGUGCCCUGCAGUUCCUGCAAAAUGUGCCCAUGGAUGAGGGUAGCAUUAGUCAAUCGGGAAUGAAGUACGAGAAGAUAA ..(((((((((.........))))(((....)))....(((((..(((((...))))).....(((((........)))))))))).)))))(((...........)))........ ( -27.60) >DroVir_CAF1 247646 117 - 1 UACUCAACGAUGUGGGCGCCGUGCAGGAGUUUCUGCAGAAUGCGCUGCAGUUCCUGCAGAACGUGCCCAUGGACGAGGGCAGCAUCAGUCAGUCGGGCAUGAAGUAUGAGAAGAUCA ..((((((...((((((((..((((((((...((((((......))))))))))))))....)))))))).(((...(((.......))).))).........)).))))....... ( -44.30) >DroGri_CAF1 260752 117 - 1 UCCUCAACGAUGUACAGGCGGUGCAGGAGUUUUUGCAGAAUGCGUUGCAAUUCCUGCAGAAUGUGCCCAUGGACGAGGGCAGCAUCAGUGAGUCGGGCAUGAAGUAUGAGAAUAUUA ..((((((.((((...(((..(((((((((...(((((......))))))))))))))((.(((((((........))))).)))).....)))..))))...)).))))....... ( -38.10) >DroMoj_CAF1 293675 117 - 1 UACUGAACGACGUGAGCGCCGUGCAGGAGUUUCUGCAGAACGCGCUGCAGUUCCUGCAGAACGUGCCCAUGGACGAGGGCAGCAUCAGUGAGUCGGGCAUGAAGUACGAGAAGAUCA ((((((..(.(....))((..((((((((...((((((......)))))))))))))).....(((((........))))))).))))))..(((..(.....)..)))........ ( -41.20) >DroAna_CAF1 181614 117 - 1 UCCUGGCGGACGUCAGUCUGAUCCAGGAGUUCCUGCAGAGCGCACUGCAGUUCCUACAAAACGUUCCUAUGGACGAGGGCAGCAUCAGCCAGUCGGGAAUGAAGUACGAAAAGAUAA (((((((((((....)))))..))))))....((((((......))))))(((.(((....(((((((...(((...(((.......))).))))))))))..))).)))....... ( -41.20) >DroPer_CAF1 282056 117 - 1 UCCUCACGGAUGUGGGAGUGGUGCAGGAAUUUCUGCAGAAUGCCCUGCAGUUUCUGCAGAACGUGCCCAUGGACGAGGGUAGCAUCAGUCAGUCAGGCAUGAAGUACGAGAAGAUCA ..(((.((..((((((.((..((((((((...((((((......))))))))))))))..))...))))))..))...(((.(.((((((.....))).))).)))))))....... ( -41.70) >consensus UCCUCAACGAUGUGAGCGCGGUGCAGGAGUUUCUGCAGAAUGCGCUGCAGUUCCUGCAGAACGUGCCCAUGGACGAGGGCAGCAUCAGUCAGUCGGGCAUGAAGUACGAGAAGAUCA ..........((((.......((((((((...((((((......))))))))))))))...(((((((...(((...(((.......))).))))))))))...))))......... (-29.28 = -28.27 + -1.02)

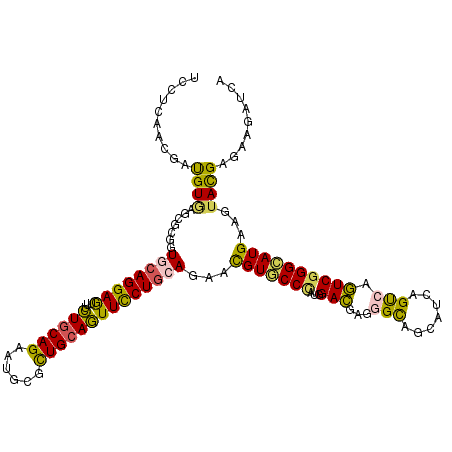

| Location | 17,974,981 – 17,975,098 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -47.15 |
| Consensus MFE | -29.02 |
| Energy contribution | -28.67 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17974981 117 - 22407834 CGGUAUUGGGAGUCUCUGGCGAAUAGCCUGCGAACUGCCAUUUUGACGGAUGUGAGUUUAAUCCAGGAAUUCCUACAGAGUGCCCUGCAGUUCCUGCAAAAUGUGCCCAUGGAUGAG .(((((.(((...(((((..((((..((((.(((((..((((......))))..)))))....)))).))))...)))))..)))(((((...)))))....))))).......... ( -34.60) >DroVir_CAF1 247686 117 - 1 CGCUAUUGGGAGGCGCUCGCGAAUAGUCUGCGUGCAUCGAUACUCAACGAUGUGGGCGCCGUGCAGGAGUUUCUGCAGAAUGCGCUGCAGUUCCUGCAGAACGUGCCCAUGGACGAG ((((((.(((.((((((((((.......)))..((((((........))))))))))))).((((((((...((((((......)))))))))))))).......))))))).)).. ( -52.30) >DroPse_CAF1 280693 117 - 1 CGGUACUGGGAGUCGCUGUCCAACAGCUUACGCUCCUCCAUCCUCACGGAUGUGGGAGUGGUGCAGGAAUUUCUGCAGAAUGCCCUGCAGUUUCUGCAGAACGUGCCCAUGGAUGAG .(((((........((((.....))))...((((((..(((((....)))))..)))))).((((((((...((((((......))))))))))))))....))))).......... ( -49.90) >DroMoj_CAF1 293715 117 - 1 CGCUACUGGGAGGCGCUCGCGACGAGCCUGCGCGCGUCCAUACUGAACGACGUGAGCGCCGUGCAGGAGUUUCUGCAGAACGCGCUGCAGUUCCUGCAGAACGUGCCCAUGGACGAG (((((.((((.(((((((((((((.((....)).))))(.....)......))))))))).((((((((...((((((......)))))))))))))).......))))))).)).. ( -54.10) >DroAna_CAF1 181654 117 - 1 CGCUACUGGGAGGCGUUGGCCAGCAGUCUCCGGUCCUCGAUCCUGGCGGACGUCAGUCUGAUCCAGGAGUUCCUGCAGAGCGCACUGCAGUUCCUACAAAACGUUCCUAUGGACGAG ........(((((((((....))).)))))).((((....(((((((((((....)))))..))))))....((((((......))))))....................))))... ( -41.10) >DroPer_CAF1 282096 117 - 1 CGGUACUGGGAGUCGCUGUCCAACAGCUUACGCUCCUCCAUCCUCACGGAUGUGGGAGUGGUGCAGGAAUUUCUGCAGAAUGCCCUGCAGUUUCUGCAGAACGUGCCCAUGGACGAG ((...(((((.(((((((.....))))...((((((..(((((....)))))..)))))).((((((((...((((((......))))))))))))))).))...)))).)..)).. ( -50.90) >consensus CGCUACUGGGAGGCGCUGGCCAACAGCCUGCGCUCCUCCAUCCUCACGGAUGUGAGAGUGGUGCAGGAAUUUCUGCAGAAUGCCCUGCAGUUCCUGCAGAACGUGCCCAUGGACGAG ((....((((....((((.....))))....((((...(((((....)))))...))))..((((((((...((((((......)))))))))))))).......))))....)).. (-29.02 = -28.67 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:08 2006