| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 240,104 – 240,206 |
| Length | 102 |
| Max. P | 0.500000 |

| Location | 240,104 – 240,206 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -16.29 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 240104 102 - 22407834 CAGGGGGUGCUCAGAAUCAGCACCAGCAGCAACCCCAACUGGCGGCCAUGUCUUCGCCGUUGCAUCAGCAAAAUCUGCCUAACAUGCACCAUUUAAUUCAA---C ..((.((((((.......)))))).((((((((.......(((((........))))))))))....(((.....)))......))).))...........---. ( -27.31) >DroPse_CAF1 38816 101 - 1 CAGCAGCAGC----AACCAACAGCAGCAACAGCCCCAAAUCUCGGCCAUGUCCUCGCCUUUGCACCAGCAAAGCCUACCCAGUAUGCAUCAUUUGAUGCAACAGC ..((.((.((----........)).))....(((.........))).........))((((((....))))))........((.((((((....))))))))... ( -22.00) >DroSim_CAF1 33435 102 - 1 CAGGAGGUGCUCAGAAUCAGCACCAGCAGCAACCCCAACUGGCGGCCAUGUCUUCGCCGUUGCAUCAGCAAAAUCUGCCUAACAUGCACCAUUUAAUUCAA---C ..((.((((((.......)))))).((((((((.......(((((........))))))))))....(((.....)))......))).))...........---. ( -27.31) >DroEre_CAF1 33389 102 - 1 CAGGAGGUGCUCAAAAUCAGCACCAACAGCAACCCCAACUGGCGGCCAUGUCUUCGCCGUUGCAUCAGCAAAAUCUGCCCAACAUGCACCAUUUAAUUCAA---C ..((.((((((.......))))))....(((.........(((((........)))))((((...(((......)))..)))).))).))...........---. ( -25.90) >DroYak_CAF1 35028 102 - 1 CAGGAGGUGCUCAGAAUCAGCACCAGCAGCAACCCCAACUGGCGGCCAUGUCUUCGCCGUUGCAUCAGCAAAACCUGCCCAACAUGCACCAUUUAAUCCAA---C ..(((((((((.......)))))).((((((((.......(((((........))))))))))....(((.....)))......))).........)))..---. ( -28.71) >DroPer_CAF1 39233 101 - 1 CAGCAGCAGC----AACCAACAGCAGCAACAGCCCCAAAUCUCGGCCAUGUCCUCGCCUUUGCACCAGCAAAGCCUACCCAGUAUGCAUCAUUUGAUGCAACAGC ..((.((.((----........)).))....(((.........))).........))((((((....))))))........((.((((((....))))))))... ( -22.00) >consensus CAGGAGGUGCUCAGAAUCAGCACCAGCAGCAACCCCAACUGGCGGCCAUGUCUUCGCCGUUGCAUCAGCAAAACCUGCCCAACAUGCACCAUUUAAUUCAA___C ..(((((((((.......)))))).(((............(((((........))))).((((....)))).............))).........)))...... (-16.29 = -17.02 + 0.73)

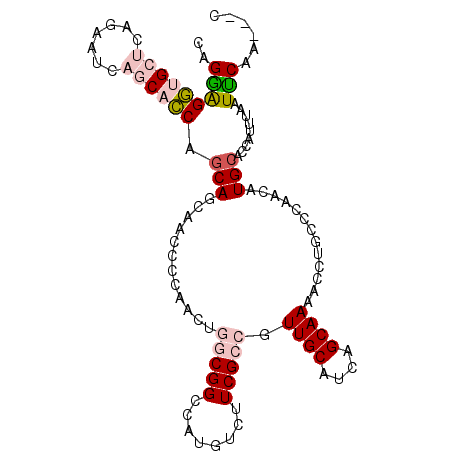
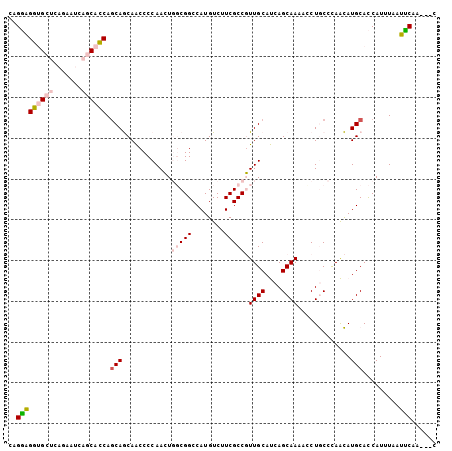
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:49 2006