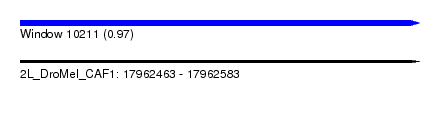
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,962,463 – 17,962,583 |
| Length | 120 |
| Max. P | 0.968172 |

| Location | 17,962,463 – 17,962,583 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -21.97 |
| Energy contribution | -22.06 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17962463 120 + 22407834 AAUGUCGCCCUCUGAUUUCGAUAGUUGCUCCAGUAGUUUAUCUUGCAUUUCCAUUCGCUGCUUCAGAAGAUGUCCUUCUUUUUGUAGCAACUCUAUCCUUUGCUGCUCCAACUCGCCAUU ..(((((...........)))))((((...(((((((.......))..........(((((...(((((.....)))))....)))))............)))))...))))........ ( -20.90) >DroVir_CAF1 233558 120 + 1 UAUAUCGCCCUCAGCCUUGGAUAGUUGCUCCAACAGUUUGUCCUGCAGUGCCAUGCGCUGCUUGAUCAGCUCACCCUCGCGCUGCAAUAGCUCAAUGCGUUGCUGCUCCAAUGCGGGAUU .......(((...((.(((((.....((......((((.(((..(((((((...)))))))..))).)))).......))((.(((((.((.....))))))).))))))).)))))... ( -38.12) >DroPse_CAF1 268564 120 + 1 AAUGUCGCCCUCCGACUUGGAGAGCUGUUCCAGCAGCUUGUCCUGCAUUUCCAUGCGCUGCUUGAGCAGAUCUCCCUCGCGCUGCAGCAGCUGGAUCCUCUGCUGCUCCAGCUCCGCAUU ...((((.....))))..(((((.((((((.((((((.......((((....)))))))))).)))))).)))))...((((((.((((((.((.....)))))))).))))...))... ( -49.41) >DroGri_CAF1 246080 120 + 1 UAUGUCUCCCUCGGCUUUGGACAGCUGCUCGAGCAGUUUAUCCUGCAACGCCAUACGCUGCUUGAGCAGCUCACCCUCGCGUUGGAGCACCUCAAUUCUCUGCUCCUCCAGCUUGGCAUU .(((((.......((...((..(((((((((((((((.(((...((...)).))).)))))))))))))))...))..))(..((((((...........))))))..).....))))). ( -43.70) >DroAna_CAF1 170100 120 + 1 GAUGUCUCCCUCUGACUUCGACAGUUGCUCCAGGAGCUUGUCUUGCAUCUCCGUCCGCUGCUUCAGCAGAUCCCCCUCCUUCUGGAGCAGUUGAAUUCUUUGCUGCUCCAGCUCUGCAUU ((.(((.......))).))((.(((.((....((((..((.....)).))))....)).))))).(((((...........((((((((((.((....)).)))))))))).)))))... ( -37.40) >DroPer_CAF1 269967 120 + 1 AAUGUCGCCCUCCGACUUGGAGAGCUGUUCCAGCAGCUUGUCCUGCAUCUCCAUGCGCUGCUUGAGAAGAUCUCCCUCGCGCUGCAGCAGCUGGAUCCUCUGCUGCUCCAGCUCCGCAUU ...((((.....))))..(((((.((.(((.((((((.......((((....)))))))))).))).)).)))))...((((((.((((((.((.....)))))))).))))...))... ( -43.11) >consensus AAUGUCGCCCUCCGACUUGGACAGCUGCUCCAGCAGCUUGUCCUGCAUCUCCAUGCGCUGCUUGAGCAGAUCUCCCUCGCGCUGCAGCAGCUCAAUCCUCUGCUGCUCCAGCUCGGCAUU .............((.(((((...((((((.((((((...................)))))).))))))..............(((((((.........)))))))))))).))...... (-21.97 = -22.06 + 0.09)

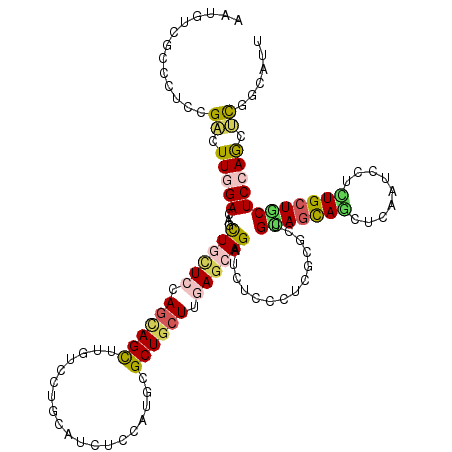

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:10:00 2006