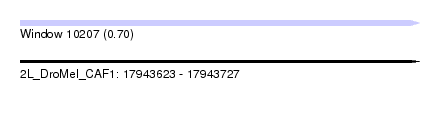
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,943,623 – 17,943,727 |
| Length | 104 |
| Max. P | 0.704348 |

| Location | 17,943,623 – 17,943,727 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -25.31 |
| Consensus MFE | -19.72 |
| Energy contribution | -18.94 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
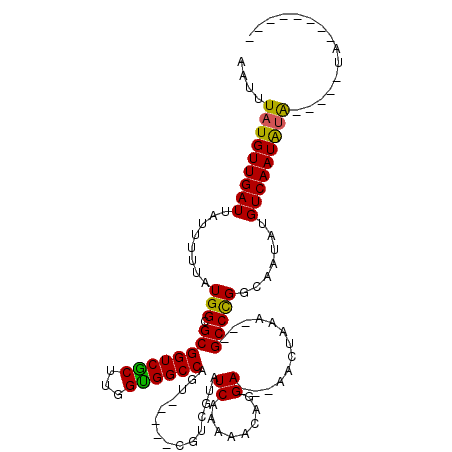
>2L_DroMel_CAF1 17943623 104 + 22407834 AAUUUAUGUUGAUUAUUUUUAUGGCGCGGUCGCUUGGUGGCCAGUCAGUCCGUCGUAUCAAAAACAGGA---AACUAAA---GCCCGGCAAUAUGUCAAUAUAUGGCAUA-------- ....(((((((((........(((.((((((((...))))))(((...(((...............)))---.)))...---))))).......))))))))).......-------- ( -24.02) >DroPse_CAF1 235526 106 + 1 AAUUUAUGUUGAUUAUUUUUAUGGCGCGGUCGCUUGGCGGCCAGU-----CGUCGUAUCAAAAGCAGGA---AACUAAA---GCCCGGCAAUAUGUCAAUAUACG-UAUACCCAUACG ....(((((((((((((..((((((((((((((...)))))).).-----)))))))......((.((.---.......---..)).)))))).)))))))))((-(((....))))) ( -29.00) >DroGri_CAF1 215799 96 + 1 AAUGUAUGUUGAUUAUUUUUAUGGCGCGGUCACUUGGUGGCCAAUCG------AGAAUCA-GAACAGGAUCCAUUUAAAUUCGCCUGGUAAUAUGUCAAUGUG--------------- ....(((((((((((((.(((.((((.((((((...)))))).....------.(.(((.-......))).).........))))))).)))).)))))))))--------------- ( -20.90) >DroMoj_CAF1 250666 96 + 1 AAUUUAUGUUGAUUAUUUUUAUGGCGCGGUCACUUGGUGGCCAAUCG------AGAAUCC-GAACAGGAUCCAUUUAAAUACGCCCGGCAAUAUGUCAAUACG--------------- ......(((((((.........((((.((((((...)))))).....------.(.((((-.....)))).).........)))).........)))))))..--------------- ( -23.87) >DroAna_CAF1 152987 95 + 1 AAUUUAUGUUGAUUAUUUUUAUGGCGCGGUCGCUUGGUGGCCAG----UCCGUCGUAUCAAAAACAGGA---AACUAAA---GCCCGGCAAUAUGUCAAUAUA-----GA-------- ..(((((((((((........(((.((((((((...))))))..----.................((..---..))...---))))).......)))))))))-----))-------- ( -25.06) >DroPer_CAF1 236585 106 + 1 AAUUUAUGUUGAUUAUUUUUAUGGCGCGGUCGCUUGGCGGCCAGU-----CGUCGUAUCAAAAGCAGGA---AACUAAA---GCCCGGCAAUAUGUCAAUAUACG-UAUACCCAUACG ....(((((((((((((..((((((((((((((...)))))).).-----)))))))......((.((.---.......---..)).)))))).)))))))))((-(((....))))) ( -29.00) >consensus AAUUUAUGUUGAUUAUUUUUAUGGCGCGGUCGCUUGGUGGCCAGU_____CGUCGUAUCAAAAACAGGA___AACUAAA___GCCCGGCAAUAUGUCAAUAUA_____UA________ ....(((((((((........(((.((((((((...))))))...............((........)).............))))).......)))))))))............... (-19.72 = -18.94 + -0.78)

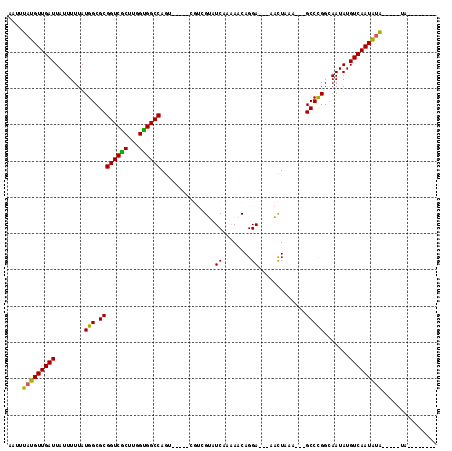
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:56 2006