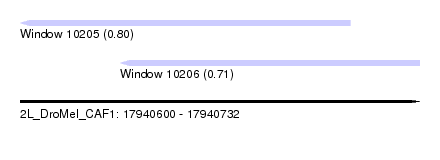
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,940,600 – 17,940,732 |
| Length | 132 |
| Max. P | 0.795545 |

| Location | 17,940,600 – 17,940,709 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.20 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -18.87 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
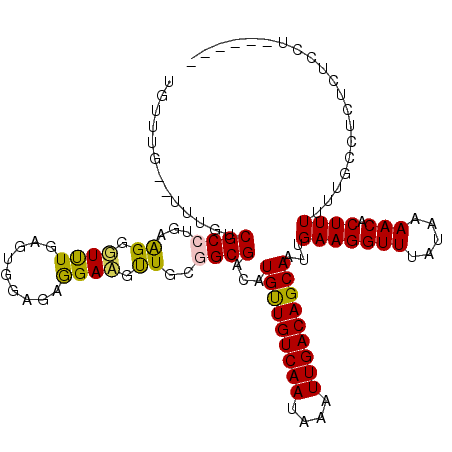
>2L_DroMel_CAF1 17940600 109 - 22407834 UAUUUU--UUGGCCGCCUGAAGGGUUUGAGUGAAAAGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUGAAGGUUUAUAAAACACUUUUUGGCCUUUUUCUU------ ......--..(((((...((((.((((..(((((.....(((((((....)....((((((....)))))))))))).....))))).)))).)))).)))))........------ ( -28.40) >DroVir_CAF1 207486 104 - 1 UGUCUG--UUUGUCGCCCGAAGCGUCUG--UG---GAGACGUUGCUGCGACAUGCUGUCAAUAAAUUGACAGCAAUUGAAGGUUUAUAAAACACUUUUGAGCCCGUGCGCU------ ......--..((((((....(((((((.--..---.)))))))...))))))(((((((((....)))))))))......((((((...........))))))........------ ( -34.60) >DroPse_CAF1 232208 109 - 1 UUUUUG--UUUGUCGCCUCAGGGUUUUGAGUGGAGAGGAAGCUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUGAAGGUUUAUAAAACACUUUUUUUCCUCACUUCU------ ......--..(((((((.(((..((((........))))..))).)))))))(((((((((....)))))))))...((((......((((.....))))......)))).------ ( -29.80) >DroMoj_CAF1 247026 104 - 1 UGUCUG--UUUGGCGCCUGAAGCGUCUG--UG---GAGACGUUGCUGCGACAUGUUGUCAAUAAAUUGACAGCAAUUGAAGGUUUAUAAAACACUUUUGAGCCCGUGAGCU------ .((((.--(..(((((.....)))))..--.)---.))))...((((((...(((((((((....)))))))))......((((((...........))))))))).))).------ ( -32.40) >DroAna_CAF1 150489 116 - 1 UCUACUCGCUUUCCGCCCGGGGGUUUUGAGUGGGGAGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUGAAGGUUUAUAAAACACUUUUUUCCCA-GCCCUCACCCAU ...(((((.(..((....))..)...)))))(((((((..((((.((.....(((((((((....)))))))))...(((((((.....))).))))...))))-)))))).))).. ( -35.90) >DroPer_CAF1 233285 109 - 1 UUUUUG--UUUGUCGCCUCAGGGUUUUGAGUGGAGAGGAAGCUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUGAAGGUUUAUAAAACACUUUUUUUCCUCACUUCU------ ......--..(((((((.(((..((((........))))..))).)))))))(((((((((....)))))))))...((((......((((.....))))......)))).------ ( -29.80) >consensus UGUUUG__UUUGUCGCCUGAAGGGUUUGAGUGGAGAGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUGAAGGUUUAUAAAACACUUUUUUGCCUCUCUCCU______ .............((((...((.((((.........)))).))..))))...(((((((((....)))))))))...(((((((.....))).)))).................... (-18.87 = -17.98 + -0.89)

| Location | 17,940,633 – 17,940,732 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -14.17 |
| Energy contribution | -13.87 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
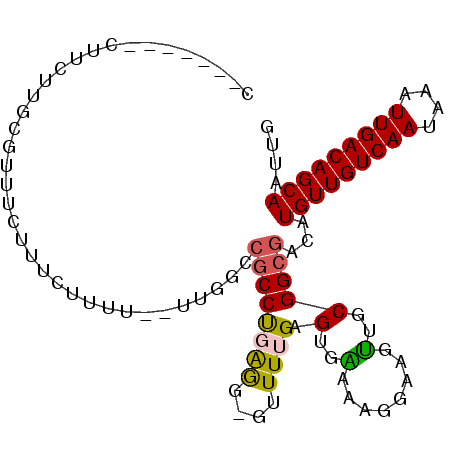
>2L_DroMel_CAF1 17940633 99 - 22407834 CCCCCCGACUACUUGCGUUUCUUUAUUUU--UUGGCCGCCUGAAG-GGUUUGAGUGAAAAGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUG ...((((...((((.(.....(((((((.--..((((........-)))).)))))))..).))))..))).)...(((((((((....))))))))).... ( -24.60) >DroPse_CAF1 232241 91 - 1 --------UUUCUUCAUAUUUUUUUUUUG--UUUGUCGCCUCAGG-GUUUUGAGUGGAGAGGAAGCUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUG --------.....................--..(((((((.(((.-.((((........))))..))).)))))))(((((((((....))))))))).... ( -27.50) >DroEre_CAF1 148113 95 - 1 CC-CCCAAGUCCUUGCGUUUCUUUCUUUU--UUGG---CCUGAAG-GCUUUGAGUGAAACGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUG .(-(((((...(((.((((((...(((..--..((---((....)-)))..))).)))))).)))))).)).)...(((((((((....))))))))).... ( -30.00) >DroYak_CAF1 156126 99 - 1 CC-CCCGACAUCUUGCGUUUCUUUCUUUU--UUGGGCGCCUGAGGCGUUUUGAGUGAAAAGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUG ..-.........((((((..(((((((((--(.(((((((...))))))).....))))))))))..)).))))..(((((((((....))))))))).... ( -32.00) >DroAna_CAF1 150527 90 - 1 G-----------UUUCGGUUCUUUCUACUCGCUUUCCGCCCGGGG-GUUUUGAGUGGGGAGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUG (-----------((.(.((.((((((.(((((((...(((....)-))...))))))).))))))..)).).))).(((((((((....))))))))).... ( -34.30) >DroPer_CAF1 233318 91 - 1 --------UUUCUUCAUAUUUUUUUUUUG--UUUGUCGCCUCAGG-GUUUUGAGUGGAGAGGAAGCUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUG --------.....................--..(((((((.(((.-.((((........))))..))).)))))))(((((((((....))))))))).... ( -27.50) >consensus C_______CUUCUUGCGUUUCUUUCUUUU__UUGGCCGCCUGAGG_GUUUUGAGUGAAAAGGAAGUUGCGGCGACAUGUUGUCAAUAAAUUGACAGCAAUUG ....................................((((((((....)))).(..(........)..)))))...(((((((((....))))))))).... (-14.17 = -13.87 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:55 2006