| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,917,175 – 1,917,280 |
| Length | 105 |
| Max. P | 0.703366 |

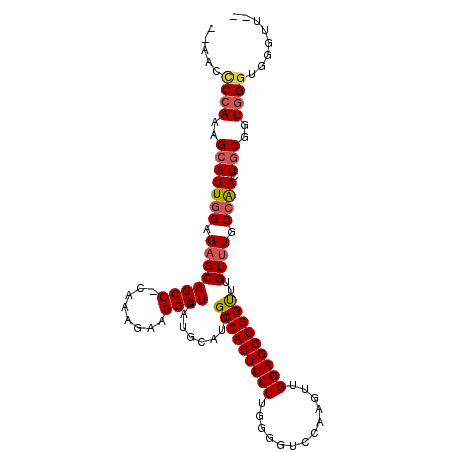
| Location | 1,917,175 – 1,917,280 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.69 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1917175 105 + 22407834 ACAACCCACCCACCGCAGUUGGCAAACAAACCACCUCCAACUUGGACCCCAAAAAGUGGCAAUGCAUUAGCCUUCUUUGGGGCUGCUCUCCAAUGCUUUGGACUU-- .........(((..((((((((..............)))))((((((((((((....(((.........)))...)))))))......))))))))..)))....-- ( -26.94) >DroSec_CAF1 82006 101 + 1 --AACCCACCCACCGCAGUUGGCAAACAAACCACCUCCAACUUGGACC-CAAAAAGUGGCAAUGCAUUAGCCUUCUUUG-GGCUGCUCUCCAAUGCUUUGGGGUU-- --(((((.....((((.(((((..............)))))(((....-)))...))))(((.((((((((((.....)-))))).......)))).))))))))-- ( -27.45) >DroSim_CAF1 65975 104 + 1 ACAACCCACCCACCGCAGUUGGCAAACAAGCCACCUCCAACUUGGACCCCAAAAAGUGGCAAUGCAUUAGCCUUCUUUG-GGCUGCUCUCCAAUGCUUUGGGGUU-- ..(((((.....((((.(.((((......)))).)(((.....))).........))))(((.((((((((((.....)-))))).......)))).))))))))-- ( -30.71) >DroEre_CAF1 82721 97 + 1 ---------CCACCGAAGUCGGCAAACAAACCACCUCCAACUUGGACUUUAAAAAGUGGCAAUGCAUUAGCCUUCUACG-GGCUGCACUCAAAUACUUUUGGUUUUU ---------...(((....))).....(((((...(((.....))).....(((((((....((((...((((.....)-)))))))......)))))))))))).. ( -21.30) >consensus __AACCCACCCACCGCAGUUGGCAAACAAACCACCUCCAACUUGGACCCCAAAAAGUGGCAAUGCAUUAGCCUUCUUUG_GGCUGCUCUCCAAUGCUUUGGGGUU__ ..............((((((((........))...(((.....)))......((((.(((.........)))..))))..)))))).((((((....)))))).... (-17.55 = -18.05 + 0.50)

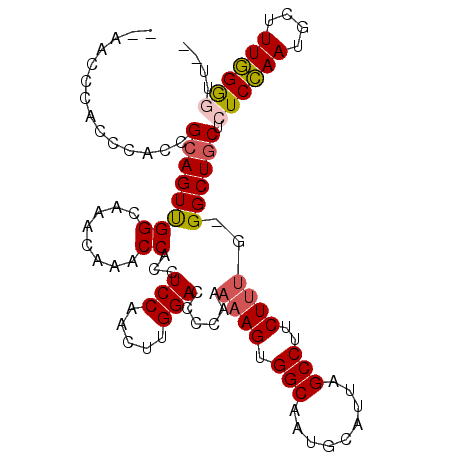
| Location | 1,917,175 – 1,917,280 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.69 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -25.27 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1917175 105 - 22407834 --AAGUCCAAAGCAUUGGAGAGCAGCCCCAAAGAAGGCUAAUGCAUUGCCACUUUUUGGGGUCCAAGUUGGAGGUGGUUUGUUUGCCAACUGCGGUGGGUGGGUUGU --...((((..(((((((..((((((((((((((.(((.........)))..)))))))))(((.....)))......)))))..)))).)))..))))........ ( -36.40) >DroSec_CAF1 82006 101 - 1 --AACCCCAAAGCAUUGGAGAGCAGCC-CAAAGAAGGCUAAUGCAUUGCCACUUUUUG-GGUCCAAGUUGGAGGUGGUUUGUUUGCCAACUGCGGUGGGUGGGUU-- --..(((((..(((((((..(((((((-((((((.(((.........)))..))))))-))(((.....)))......)))))..)))).)))..)))).)....-- ( -35.90) >DroSim_CAF1 65975 104 - 1 --AACCCCAAAGCAUUGGAGAGCAGCC-CAAAGAAGGCUAAUGCAUUGCCACUUUUUGGGGUCCAAGUUGGAGGUGGCUUGUUUGCCAACUGCGGUGGGUGGGUUGU --..(((((..(((((((..(((((((-((((((.(((.........)))..)))))))).(((.....)))......)))))..)))).)))..)))).)...... ( -36.50) >DroEre_CAF1 82721 97 - 1 AAAAACCAAAAGUAUUUGAGUGCAGCC-CGUAGAAGGCUAAUGCAUUGCCACUUUUUAAAGUCCAAGUUGGAGGUGGUUUGUUUGCCGACUUCGGUGG--------- .....(((...((((....))))...(-((.((..(((....(((..(((((((((.............))))))))).)))..)))..)).))))))--------- ( -26.32) >consensus __AACCCCAAAGCAUUGGAGAGCAGCC_CAAAGAAGGCUAAUGCAUUGCCACUUUUUGGGGUCCAAGUUGGAGGUGGUUUGUUUGCCAACUGCGGUGGGUGGGUU__ .....((((..(((((((.((((((((........))))........(((((((((.............)))))))))..)))).)))).)))..))))........ (-25.27 = -26.02 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:38 2006