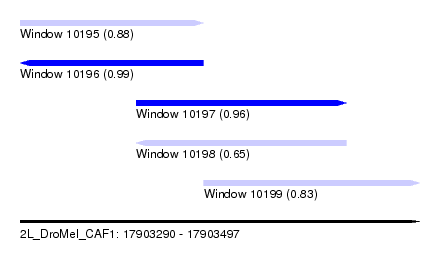
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,903,290 – 17,903,497 |
| Length | 207 |
| Max. P | 0.994057 |

| Location | 17,903,290 – 17,903,385 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
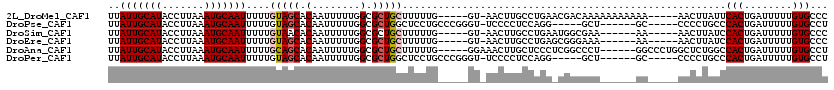
>2L_DroMel_CAF1 17903290 95 + 22407834 AAAGGCUGCUGUUUGGGCCAACGGCAGUUGCAGUGCAAGCG-AAAAAAUUGGCCAGCAGAAGGGCACAAAAAUCAGUGAAUAAGUUUUUUUUUUUU (((((((.((((...((((((......((((.......)))-).....)))))).)))).....(((........)))....)))))))....... ( -21.50) >DroSec_CAF1 123143 90 + 1 AAAGGCUGCUGUUUGGGCCAACGGCAGUCGCAGUGCAAGCGGAAAAAAUUGGCCAGCAGAAGGGCACAAAAAUCAGUGGAUAAGUUUU------UU ((((((((((.(((.((((((......((((.......))))......))))))....))).)))......(((....))).))))))------). ( -23.70) >DroSim_CAF1 100867 90 + 1 AAAGGCUGCUGUUUGGGCCAACGGCAGUCGCAGUGCAAGCGGAAAAAAUUGGCCAGCAGAAGGGCACAAAAAUCAGUGGAUAAGUUUU------UU ((((((((((.(((.((((((......((((.......))))......))))))....))).)))......(((....))).))))))------). ( -23.70) >DroEre_CAF1 112887 89 + 1 AAAGGCUGCUCUUUGGGCCAAUGGCAGUCGCAGUGGAAGCG-AAAAAAUUGGCCAGCAGAAGGGCACAAAAAUCAGUGGAUAAGUUUU------UU ((((((((((((((.(((((((.....((((.......)))-)....)))))))....)))))))......(((....))).))))))------). ( -29.80) >DroYak_CAF1 120437 89 + 1 AAAGGCUGCUGUUUGGGCCAACGGCAGUCGCAGUGGAAGCG-AAAAAAUUGGCCCGCAGAAGGGCACAAAAAUCAGUGGAUAAGUUUU------UU ...((((((((((......)))))))))).....((((((.-......(((((((......)))).)))..(((....)))..)))))------). ( -27.40) >consensus AAAGGCUGCUGUUUGGGCCAACGGCAGUCGCAGUGCAAGCG_AAAAAAUUGGCCAGCAGAAGGGCACAAAAAUCAGUGGAUAAGUUUU______UU ...((((((((((......))))))))))((.......))........((((((........))).)))........................... (-20.40 = -20.44 + 0.04)


| Location | 17,903,290 – 17,903,385 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17903290 95 - 22407834 AAAAAAAAAAAACUUAUUCACUGAUUUUUGUGCCCUUCUGCUGGCCAAUUUUUU-CGCUUGCACUGCAACUGCCGUUGGCCCAAACAGCAGCCUUU ..................(((........))).....((((((((((((.....-((.((((...)))).))..))))))).....)))))..... ( -17.30) >DroSec_CAF1 123143 90 - 1 AA------AAAACUUAUCCACUGAUUUUUGUGCCCUUCUGCUGGCCAAUUUUUUCCGCUUGCACUGCGACUGCCGUUGGCCCAAACAGCAGCCUUU ..------..........(((........))).....((((((((((((......(((.......)))......))))))).....)))))..... ( -20.00) >DroSim_CAF1 100867 90 - 1 AA------AAAACUUAUCCACUGAUUUUUGUGCCCUUCUGCUGGCCAAUUUUUUCCGCUUGCACUGCGACUGCCGUUGGCCCAAACAGCAGCCUUU ..------..........(((........))).....((((((((((((......(((.......)))......))))))).....)))))..... ( -20.00) >DroEre_CAF1 112887 89 - 1 AA------AAAACUUAUCCACUGAUUUUUGUGCCCUUCUGCUGGCCAAUUUUUU-CGCUUCCACUGCGACUGCCAUUGGCCCAAAGAGCAGCCUUU ..------..........(((........))).....((((((((((((....(-(((.......)))).....))))))).....)))))..... ( -21.20) >DroYak_CAF1 120437 89 - 1 AA------AAAACUUAUCCACUGAUUUUUGUGCCCUUCUGCGGGCCAAUUUUUU-CGCUUCCACUGCGACUGCCGUUGGCCCAAACAGCAGCCUUU ..------..........(((........))).....((((((((((((....(-(((.......)))).....)))))))).....))))..... ( -23.20) >consensus AA______AAAACUUAUCCACUGAUUUUUGUGCCCUUCUGCUGGCCAAUUUUUU_CGCUUGCACUGCGACUGCCGUUGGCCCAAACAGCAGCCUUU ..................(((........))).....((((((((((((......(((.......)))......))))))).....)))))..... (-18.58 = -18.82 + 0.24)


| Location | 17,903,350 – 17,903,459 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.29 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -12.91 |
| Energy contribution | -12.99 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17903350 109 + 22407834 GGGCACAAAAAUCAGUGAAUAAGUU-----UUUUUUUUUUUGUCGUUCAGGCAAGUU-AC-----CAAAAAGCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAA .(((((((......(((.....(((-----((((....((((((.....))))))..-..-----.)))))))..)))......)))))))......(((((((.......))))))).. ( -24.70) >DroPse_CAF1 194820 103 + 1 AGGCACAAAAAUCAGUGGGCAGGGG-----GC------AGC-----CCUGGAGGGGA-ACCCGGGCAGGAGCCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAA .(((((((.......(((((..(((-----..------..(-----((.....))).-.))).(((....))).)).)))....)))))))......(((((((.......))))))).. ( -35.20) >DroSim_CAF1 100928 103 + 1 GGGCACAAAAAUCAGUGGAUAAGUU-----UU------UUCGCCAUUCAGGCAAGUU-AC-----CAAAAAGCAGCGCCAAAAAUUGUGUUACAAAAAUUGCAUUUAAGGUAUGCAAUAA .(((((((.......(((....(((-----((------((.(((.....)))..(..-..-----))))))))....)))....)))))))......(((((((.......))))))).. ( -20.70) >DroEre_CAF1 112947 103 + 1 GGGCACAAAAAUCAGUGGAUAAGUU-----UU------UUUCCCGCUCAGGCAAGUU-AC-----CAAAAAGCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAA .(((((((.....(((((.......-----..------....)))))..(((..(((-..-----.....)))...))).....)))))))......(((((((.......))))))).. ( -23.42) >DroAna_CAF1 116873 109 + 1 AGGCACAAAAAUCAGUGGCCAGAGCCAGGGCC------AGGGCCGAGGGAGCAAGUUUCC-----CAAAAAGCAGCGCCAAAAAUUGUGCUGCAAAAAUUGCAUUUAAGGUAUGCAAUAA .(((((........))((((........))))------...)))..(((((.....))))-----).....(((((((........)))))))....(((((((.......))))))).. ( -36.00) >DroPer_CAF1 195930 103 + 1 AGGCACAAAAAUCAGUGGGCAGGGG-----GC------AGC-----CCUGGAGGGGA-ACCCGGGCAGGAGCCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAA .(((((((.......(((((..(((-----..------..(-----((.....))).-.))).(((....))).)).)))....)))))))......(((((((.......))))))).. ( -35.20) >consensus AGGCACAAAAAUCAGUGGACAAGGU_____UC______AGCGCCG_UCAGGCAAGUU_AC_____CAAAAAGCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAA .(((((((...((....))..............................(((..((...............))...))).....)))))))......(((((((.......))))))).. (-12.91 = -12.99 + 0.08)


| Location | 17,903,350 – 17,903,459 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.29 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17903350 109 - 22407834 UUAUUGCAUACCUUAAAUGCAAUUUUUGUAGCACAAUUUUUGGCGCUGCUUUUUG-----GU-AACUUGCCUGAACGACAAAAAAAAAAA-----AACUUAUUCACUGAUUUUUGUGCCC ..(((((((.......))))))).......((((((.....((((.((((....)-----))-)...))))((((..(............-----...)..)))).......)))))).. ( -17.56) >DroPse_CAF1 194820 103 - 1 UUAUUGCAUACCUUAAAUGCAAUUUUUGUAGCACAAUUUUUGGCGCUGGCUCCUGCCCGGGU-UCCCCUCCAGG-----GCU------GC-----CCCCUGCCCACUGAUUUUUGUGCCU ..(((((((.......))))))).......((((((.((.(((.((.(((....))).((((-..(((....))-----)..------))-----))...)))))..))...)))))).. ( -33.30) >DroSim_CAF1 100928 103 - 1 UUAUUGCAUACCUUAAAUGCAAUUUUUGUAACACAAUUUUUGGCGCUGCUUUUUG-----GU-AACUUGCCUGAAUGGCGAA------AA-----AACUUAUCCACUGAUUUUUGUGCCC ..(((((((.......)))))))..................(((((((((....)-----))-)..(((((.....))))).------..-----...................))))). ( -20.50) >DroEre_CAF1 112947 103 - 1 UUAUUGCAUACCUUAAAUGCAAUUUUUGUAGCACAAUUUUUGGCGCUGCUUUUUG-----GU-AACUUGCCUGAGCGGGAAA------AA-----AACUUAUCCACUGAUUUUUGUGCCC ..(((((((.......))))))).......(((((((((((..(.((((((...(-----((-.....))).)))))))..)------))-----))...(((....)))..)))))).. ( -23.70) >DroAna_CAF1 116873 109 - 1 UUAUUGCAUACCUUAAAUGCAAUUUUUGCAGCACAAUUUUUGGCGCUGCUUUUUG-----GGAAACUUGCUCCCUCGGCCCU------GGCCCUGGCUCUGGCCACUGAUUUUUGUGCCU ..(((((((.......))))))).......((((((...(..(...........(-----(((.......))))..((((..------(((....)))..)))).)..)...)))))).. ( -32.40) >DroPer_CAF1 195930 103 - 1 UUAUUGCAUACCUUAAAUGCAAUUUUUGUAGCACAAUUUUUGGCGCUGGCUCCUGCCCGGGU-UCCCCUCCAGG-----GCU------GC-----CCCCUGCCCACUGAUUUUUGUGCCU ..(((((((.......))))))).......((((((.((.(((.((.(((....))).((((-..(((....))-----)..------))-----))...)))))..))...)))))).. ( -33.30) >consensus UUAUUGCAUACCUUAAAUGCAAUUUUUGUAGCACAAUUUUUGGCGCUGCUUUUUG_____GU_AACUUGCCUGA_CGGCGAA______AA_____AACCUAUCCACUGAUUUUUGUGCCC ..(((((((.......)))))))....(((((.(........).)))))......................................................(((........)))... (-10.52 = -10.88 + 0.36)


| Location | 17,903,385 – 17,903,497 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -19.03 |
| Energy contribution | -20.12 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17903385 112 + 22407834 UGUCGUUCAGGCAAGUU-AC-----CAAAAAGCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAAAAUGCCAUGCAUA--UUUGGCUCUUGCUGUGCUUUUUGCA ((((.....))))....-..-----(((((((((((((........)))))(((...(((((((.......))))))).....((((......--..))))......))))))))))).. ( -29.20) >DroSim_CAF1 100957 112 + 1 CGCCAUUCAGGCAAGUU-AC-----CAAAAAGCAGCGCCAAAAAUUGUGUUACAAAAAUUGCAUUUAAGGUAUGCAAUAAAAUGCCAUGCAUA--UUUGGCUCUUGCUGUGCUUUUUGCA .(((.....))).....-..-----(((((((((((((((((..(((.....)))....(((((....(((((........))))))))))..--))))))....))..))))))))).. ( -30.20) >DroEre_CAF1 112976 112 + 1 UCCCGCUCAGGCAAGUU-AC-----CAAAAAGCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAAAAUGCCAUGCAUA--UUUUGGUCUUGCUGGGCUUUUUGCC ....((((((.((((..-((-----(((((.(.(((((........))))).)......(((((....(((((........))))))))))..--)))))))))))))))))........ ( -34.40) >DroYak_CAF1 120526 111 + 1 UCCCGUUCAUGCAAGUU-AC-----CAAAAAGCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAAAAUGCCAUGCAUA--UU-UGGUCUUGCUGGGCUUUUUGCA ....(((((.(((((..-((-----((((..(.(((((........))))).)......(((((....(((((........))))))))))..--))-))))))))))))))........ ( -31.00) >DroAna_CAF1 116907 115 + 1 GGCCGAGGGAGCAAGUUUCC-----CAAAAAGCAGCGCCAAAAAUUGUGCUGCAAAAAUUGCAUUUAAGGUAUGCAAUAAAAUGCCAUGCAUACAUUUUGCUCUUCUUGCUCUUUUUGCC (((.(((((((((((.....-----(((((.(((((((........)))))))......(((((....(((((........))))))))))....))))).....))))))))))).))) ( -42.70) >DroPer_CAF1 195959 104 + 1 C-----CCUGGAGGGGA-ACCCGGGCAGGAGCCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAAAAUGCCAUGCAUACAUUUUGCUCUU-------UUU---UU .-----(((((.(....-.)))))).((((((.(((((........)))))........(((((....(((((........))))))))))........))))))-------...---.. ( -27.50) >consensus UGCCGUUCAGGCAAGUU_AC_____CAAAAAGCAGCGCCAAAAAUUGUGCUACAAAAAUUGCAUUUAAGGUAUGCAAUAAAAUGCCAUGCAUA__UUUUGCUCUUGCUGGGCUUUUUGCA .........................(((((((((((((........))))).(((((..(((((....(((((........))))))))))....)))))..........)))))))).. (-19.03 = -20.12 + 1.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:48 2006