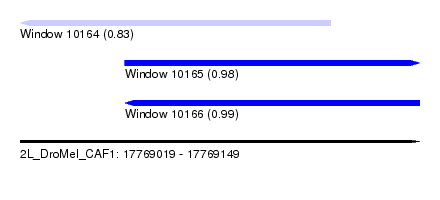
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,769,019 – 17,769,149 |
| Length | 130 |
| Max. P | 0.993734 |

| Location | 17,769,019 – 17,769,120 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.34 |
| Mean single sequence MFE | -15.46 |
| Consensus MFE | -8.47 |
| Energy contribution | -9.08 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
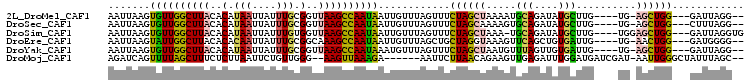
>2L_DroMel_CAF1 17769019 101 - 22407834 UUUUAGCUAGAAACUAAACAAUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUUACCCAUUCAA-----AAUCAGAGACCCUGCACUUUG-CCA .(((((.......)))))......((((((((..((((........))))))))))))...................-----...(((((........)))))-... ( -14.40) >DroSec_CAF1 68931 94 - 1 CUUUUGCUAGAAACUAAACAAUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUUACCCAUUCAA-----AAUCAGAGA-------CUUUG-CCA .....((.((..............((((((((..((((........))))))))))))..............(((..-----.....))).-------))..)-).. ( -12.20) >DroSim_CAF1 72804 99 - 1 -UUUAGCUAGAAACUAAACAAUUAUUGGCUUAACCACAAAUAAUUAUGUGUAAGCCAACACUUAAUUACCCAU-CAA-----AAUCAGAGACCCAGCACUUUG-CCA -(((((.......)))))......((((((((..((((........))))))))))))...............-...-----...(((((........)))))-... ( -14.60) >DroEre_CAF1 78382 100 - 1 CUUUACCUAGCAGCUAAACAAUUAUUGGCUUUGCCGCAAAUAAUUGUGUGUAAGCCAAUACUUAAUUACCCAUUCAA-----AAUCAGAGAC-UGGCACUCUG-CCA .........((((.........(((((((((.(((((((....))))).))))))))))).........(((.((..-----.......)).-)))....)))-).. ( -22.50) >DroYak_CAF1 71273 100 - 1 CAUUAGCUAGAAACUAAACAUUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUUACCCAUUCAA-----AAUCAGAUAC-UGGCACUAUG-CCA .....(((((..............((((((((..((((........))))))))))))...............((..-----.....))..)-))))......-... ( -17.50) >DroMoj_CAF1 105249 88 - 1 CUUCUGUUAAGAAUU------UCUUUAACUU--CCCAACAGAAUUAAGAGAAAGCUAAAACUGAUCUAU----UAAGAUAUUAAACAGAUA-------UUUUCUUCA .(((((((..(((..------........))--)..)))))))..(((((((..((.......((((..----..)))).......))...-------))))))).. ( -11.54) >consensus CUUUAGCUAGAAACUAAACAAUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUUACCCAUUCAA_____AAUCAGAGAC___GCACUUUG_CCA ........................((((((((..((((........))))))))))))................................................. ( -8.47 = -9.08 + 0.61)

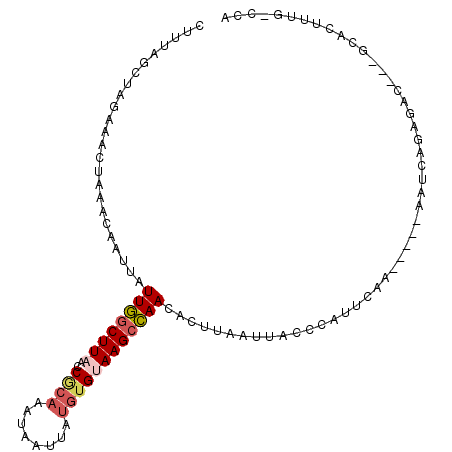
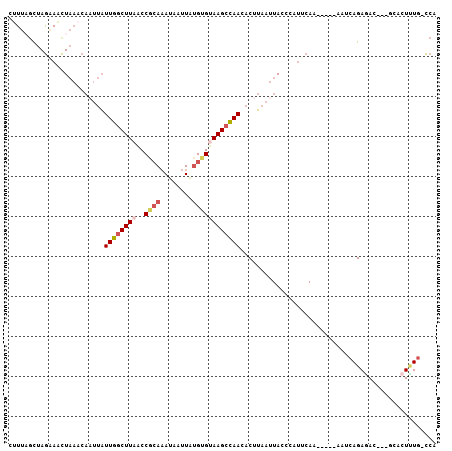
| Location | 17,769,053 – 17,769,149 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.50 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -12.43 |
| Energy contribution | -11.79 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17769053 96 + 22407834 AAUUAAGUGUUGGCUUACACAUAAUUAUUUGCGGUUAAGCCAAUAAUUGUUUAGUUUCUAGCUAAAAUGCAGAUAUGCUUG----UG-AGCUGG---GAUUAGG-- .....(((.(..(((((((......(((((((((.....))........((((((.....))))))..)))))))....))----))-)))..)---.)))...-- ( -26.30) >DroSec_CAF1 68958 96 + 1 AAUUAAGUGUUGGCUUACACAUAAUUAUUUGCGGUUAAGCCAAUAAUUGUUUAGUUUCUAGCAAAAGUGCAGAUAUGCUUG----UG-AGCUGG---CUUUAGG-- ..(((((.((..(((((((.(((((((((.((......)).)))))))))..(((.(((.(((....))))))...)))))----))-)))..)---)))))).-- ( -28.30) >DroSim_CAF1 72837 98 + 1 AAUUAAGUGUUGGCUUACACAUAAUUAUUUGUGGUUAAGCCAAUAAUUGUUUAGUUUCUAGCUAAA-UGCAGAUAUGCUUG----UGGAGCUGG---GAUUAGGUG .......((((((((((..(((((....)))))..))))))))))...(..(((((.((((((...-.(((....)))...----...))))))---)))))..). ( -30.10) >DroEre_CAF1 78415 96 + 1 AAUUAAGUAUUGGCUUACACACAAUUAUUUGCGGCAAAGCCAAUAAUUGUUUAGCUGCUAGGUAAAGUUCAGCUGUGAUUG----UG-AACUGG---GAUGGGG-- ...(((((....)))))..(((((((((..(((((.((((........)))).)))))..(((........))))))))))----))-......---.......-- ( -20.50) >DroYak_CAF1 71306 96 + 1 AAUUAAGUGUUGGCUUACACAUAAUUAUUUGCGGUUAAGCCAAUAAAUGUUUAGUUUCUAGCUAAUGUUUAGUUGUGAUUG----UG-AGCUGG---GAUUAGG-- .....(((.(..(((((((((((((((...((((.....)).........(((((.....))))).)).))))))))..))----))-)))..)---.)))...-- ( -24.80) >DroMoj_CAF1 105278 95 + 1 AGAUCAGUUUUAGCUUUCUCUUAAUUCUGUUGGG--AAGUUAAAGA------AAUUCUUAACAGAAGUUGAGAUUUGGAUGAUCGAU-AAUUGGGCUAUUUAGC-- .(((((..(((((...((((....((((((((((--((........------..))))))))))))...)))).))))))))))...-......(((....)))-- ( -23.90) >consensus AAUUAAGUGUUGGCUUACACAUAAUUAUUUGCGGUUAAGCCAAUAAUUGUUUAGUUUCUAGCUAAAGUGCAGAUAUGCUUG____UG_AGCUGG___GAUUAGG__ .......((((((((((..(.(((....))).)..))))))))))............((((((.....(((....)))..........))))))............ (-12.43 = -11.79 + -0.63)
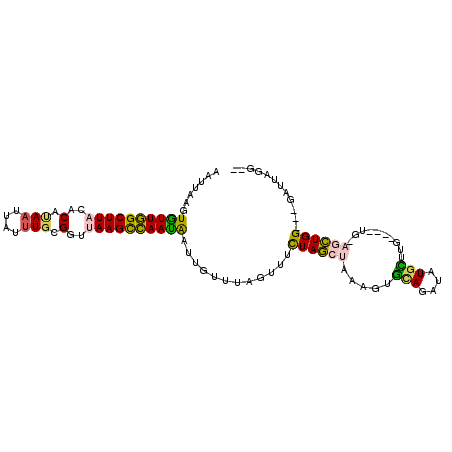
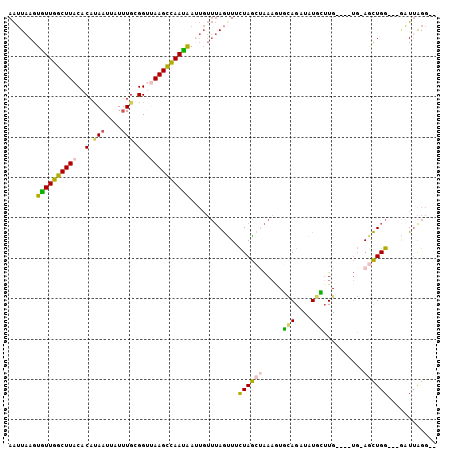
| Location | 17,769,053 – 17,769,149 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.50 |
| Mean single sequence MFE | -16.53 |
| Consensus MFE | -8.47 |
| Energy contribution | -9.08 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17769053 96 - 22407834 --CCUAAUC---CCAGCU-CA----CAAGCAUAUCUGCAUUUUAGCUAGAAACUAAACAAUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUU --.......---..((((-..----...(((....))).....))))................((((((((..((((........))))))))))))......... ( -16.80) >DroSec_CAF1 68958 96 - 1 --CCUAAAG---CCAGCU-CA----CAAGCAUAUCUGCACUUUUGCUAGAAACUAAACAAUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUU --....(((---((((((-..----..)))...((((((....))).)))..............))))))..........(((((.((((......)))).))))) ( -18.10) >DroSim_CAF1 72837 98 - 1 CACCUAAUC---CCAGCUCCA----CAAGCAUAUCUGCA-UUUAGCUAGAAACUAAACAAUUAUUGGCUUAACCACAAAUAAUUAUGUGUAAGCCAACACUUAAUU .........---...(((...----..)))...(((((.-....)).))).............((((((((..((((........))))))))))))......... ( -16.80) >DroEre_CAF1 78415 96 - 1 --CCCCAUC---CCAGUU-CA----CAAUCACAGCUGAACUUUACCUAGCAGCUAAACAAUUAUUGGCUUUGCCGCAAAUAAUUGUGUGUAAGCCAAUACUUAAUU --.......---......-..----.......(((((..((......))))))).......(((((((((.(((((((....))))).)))))))))))....... ( -21.00) >DroYak_CAF1 71306 96 - 1 --CCUAAUC---CCAGCU-CA----CAAUCACAACUAAACAUUAGCUAGAAACUAAACAUUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUU --.......---..((((-..----..................))))................((((((((..((((........))))))))))))......... ( -12.55) >DroMoj_CAF1 105278 95 - 1 --GCUAAAUAGCCCAAUU-AUCGAUCAUCCAAAUCUCAACUUCUGUUAAGAAUU------UCUUUAACUU--CCCAACAGAAUUAAGAGAAAGCUAAAACUGAUCU --(((....)))......-...((((.......((((...(((((((..(((..------........))--)..)))))))....)))).((......)))))). ( -13.90) >consensus __CCUAAUC___CCAGCU_CA____CAAGCAUAUCUGAACUUUAGCUAGAAACUAAACAAUUAUUGGCUUAACCGCAAAUAAUUAUGUGUAAGCCAACACUUAAUU ...............................................................((((((((..((((........))))))))))))......... ( -8.47 = -9.08 + 0.61)

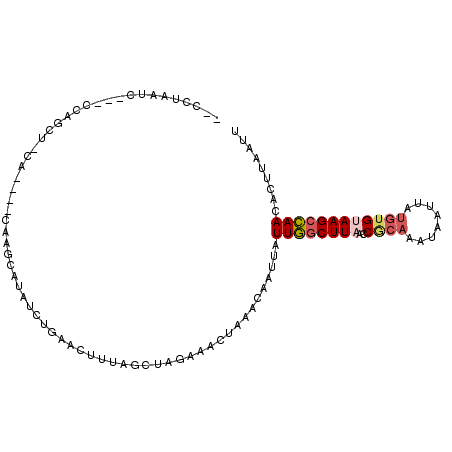
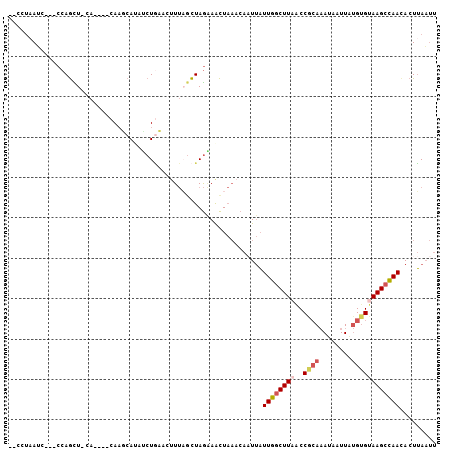
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:15 2006