| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,897,214 – 1,897,421 |
| Length | 207 |
| Max. P | 0.927550 |

| Location | 1,897,214 – 1,897,321 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.74 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -27.84 |
| Energy contribution | -27.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1897214 107 + 22407834 UUAAAACUAAACUGUGCCGGCAGUUUGGAAAUUUCACACAUUGCGGCUAAAGAGUUUUCUCAGUUGGCAGUGGCAAAUUACAGUUGCAAACAAUGCGACGAUGC-------------CAC ......(((((((((....))))))))).................(((((.(((....)))..))))).((((((.......((((((.....))))))..)))-------------))) ( -31.10) >DroEre_CAF1 62104 111 + 1 -UAUAACUAAACUGUGCCGGCAGUUUGGAAAUUUCACACAUUCCGGCUAAAGAGUUUUCUCAGUUGGCAGUGGCAAAUUACAGUUGCAAACAAUGCGACGAUGCCUUGC--------CAC -.....(((((((((....))))))))).................(((((.(((....)))..))))).(((((((..((..((((((.....))))))..))..))))--------))) ( -34.40) >DroYak_CAF1 71431 119 + 1 -UAUAACUAAACUGCGCCGGCAGUUUGGAAAUUUCACACAUUCCGGCUAAAGAGUUUUCUCAGUUGGCAGUGGCAAAUUACAGUUGCGAACAAUGCGACGAUGCCUUGCCUCAUGGCCAC -.....(((((((((....)))))))))................(((((..(((....)))....(((((.((((.......((((((.....))))))..)))))))))...))))).. ( -39.50) >consensus _UAUAACUAAACUGUGCCGGCAGUUUGGAAAUUUCACACAUUCCGGCUAAAGAGUUUUCUCAGUUGGCAGUGGCAAAUUACAGUUGCAAACAAUGCGACGAUGCCUUGC________CAC ......(((((((((....))))))))).(((((..(((...((((((...(((....)))))))))..)))..)))))...((((((.....))))))..................... (-27.84 = -27.73 + -0.11)

| Location | 1,897,294 – 1,897,384 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.38 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1897294 90 + 22407834 CAGUUGCAAACAAUGCGACGAUGC-------------CACAUGG--------CCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCA------GCUACAGUUACACG .(((((((....(((((..(((((-------------(...(((--------(.((.(((....))).))))))..)))))).....))))).))))------)))........... ( -28.30) >DroEre_CAF1 62183 101 + 1 CAGUUGCAAACAAUGCGACGAUGCCUUGC--------CACAUGGCCAAAUGCCCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCA--------CACAAAUACACG ..((((((.....))))))((((((((((--------(....)))........(((.(((....))).)))...)))))))).....(((....)))--------............ ( -27.60) >DroYak_CAF1 71510 109 + 1 CAGUUGCGAACAAUGCGACGAUGCCUUGCCUCAUGGCCACAUGGCCAAAUGGCCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCA--------CACAAAUACACG ..((((((....(((((..(((((((((((...(((((....)))))...)))(((.(((....))).)))...)))))))).....))))).))))--------.))......... ( -36.70) >DroAna_CAF1 65844 108 + 1 CAGUUAUAAACAAUUCUGCC-UGCCUUGC--------CACAUGCCCCGUCCAACAGUUGCAGUUGCAAUUGUCAGAGGAAUUAAUAAUGCAUGUGCUGGCCAGGCAACAGACACACA ..............((((..-(((((.((--------(((((((.((.((..((((((((....))))))))..))))..........))))))...))).))))).))))...... ( -33.80) >consensus CAGUUGCAAACAAUGCGACGAUGCCUUGC________CACAUGGCCAAAUG_CCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCA________CACAAAUACACG ..((((((.....))))))..................(((((...........(((.(((....))).)))......((((.....)))))))))...................... (-13.44 = -13.87 + 0.44)

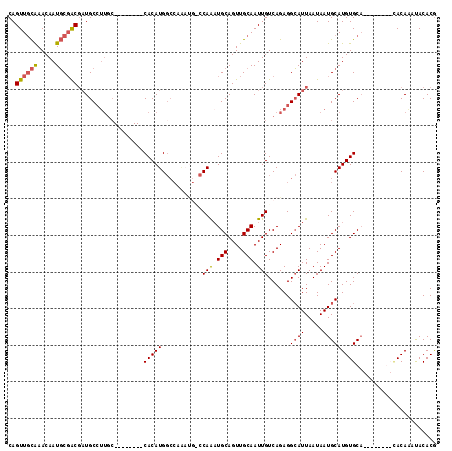
| Location | 1,897,294 – 1,897,384 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.38 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1897294 90 - 22407834 CGUGUAACUGUAGC------UGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGG--------CCAUGUG-------------GCAUCGUCGCAUUGUUUGCAACUG .(((((.(....).------))))).((((....((((((..(...(((.(((....))).)))(--------((....)-------------))...)..))))))..)))).... ( -25.40) >DroEre_CAF1 62183 101 - 1 CGUGUAUUUGUG--------UGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGGGCAUUUGGCCAUGUG--------GCAAGGCAUCGUCGCAUUGUUUGCAACUG .((((((....)--------))))).((((....((((((....(((...(((...(((.....(((.....))).....--------)))..)))..)))))))))..)))).... ( -30.10) >DroYak_CAF1 71510 109 - 1 CGUGUAUUUGUG--------UGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGGCCAUUUGGCCAUGUGGCCAUGAGGCAAGGCAUCGUCGCAUUGUUCGCAACUG .......(((((--------.(((.((((.......((((((....(((.(((....))).)))(((...(((((....)))))...))).))))))....))))))).)))))... ( -36.80) >DroAna_CAF1 65844 108 - 1 UGUGUGUCUGUUGCCUGGCCAGCACAUGCAUUAUUAAUUCCUCUGACAAUUGCAACUGCAACUGUUGGACGGGGCAUGUG--------GCAAGGCA-GGCAGAAUUGUUUAUAACUG ......((((((((((.(((...((((((.........(((((..(((.((((....)))).)))..)).))))))))))--------)).)))))-.))))).............. ( -39.00) >consensus CGUGUAUCUGUG________UGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGG_CAUUUGGCCAUGUG________GCAAGGCAUCGUCGCAUUGUUUGCAACUG .((((((.(((((......))))).))))))...((((((..(.......(((....)))..(((((....)))))......................)..)))))).......... (-15.65 = -15.40 + -0.25)

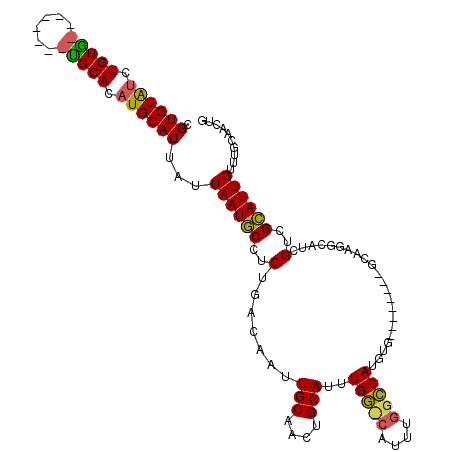
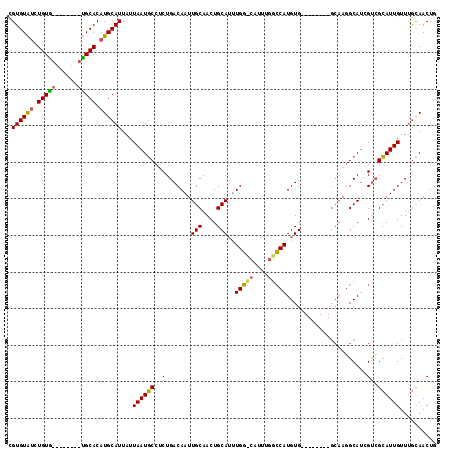
| Location | 1,897,321 – 1,897,421 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1897321 100 + 22407834 AUGG--------CCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCAGCUACAGUUACACGGCUAGAAAAAAAUGGGAAAUAC---UAAAGCCGUUGACGG ....--------((...((.(((((((.....((...((((.....)))).))))))))).))((((.((((((.........(((......)---)).)))))))))))) ( -23.10) >DroEre_CAF1 62215 108 + 1 AUGGCCAAAUGCCCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCA--CACAAAUACACGGCUAGAAAA-AAUGGAAAUAAAAAAAACAGCAGUCGACGA .(((((.(((((((((.(((....))).))).....)))))).....(((....)))--...........))))).....-.............................. ( -20.10) >DroYak_CAF1 71550 108 + 1 AUGGCCAAAUGGCCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCA--CACAAAUACACGGCUAGAAAA-AAUGGCAAAUAAACACACAGCCGUCGACGG .(((((....)))))..(((....)))..((((.(((((........(((....)))--............((((.....-..)))).............))).)))))). ( -26.40) >consensus AUGGCCAAAUG_CCAAAUGCAGUUGCAAUUGUCAGAGGCAUUAAUAAUGCAUGUGCA__CACAAAUACACGGCUAGAAAA_AAUGGAAAAUAAA_AAACAGCCGUCGACGG .(((((....)))))..(((....))).(((((....((((.....)))).((((....)))).....((((((.........................)))))).))))) (-18.05 = -18.44 + 0.39)

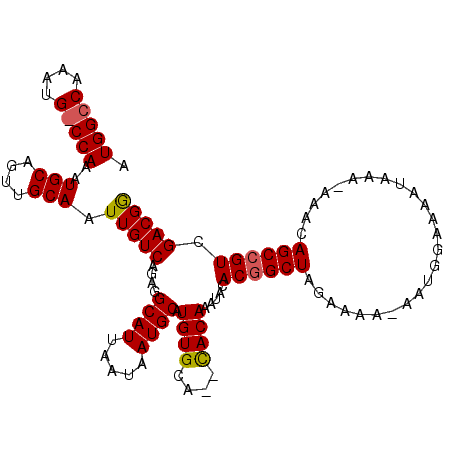
| Location | 1,897,321 – 1,897,421 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -17.69 |
| Energy contribution | -18.97 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1897321 100 - 22407834 CCGUCAACGGCUUUA---GUAUUUCCCAUUUUUUUCUAGCCGUGUAACUGUAGCUGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGG--------CCAU ..((((((((((...---...................)))))).....(((((.((((((.((((.....))))...)).....)))).)))))..)))--------)... ( -20.35) >DroEre_CAF1 62215 108 - 1 UCGUCGACUGCUGUUUUUUUUAUUUCCAUU-UUUUCUAGCCGUGUAUUUGUG--UGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGGGCAUUUGGCCAU ..(((((.((((..................-..........((((((....)--)))))..((((.....))))......(((.(((....))).))))))).)))))... ( -23.10) >DroYak_CAF1 71550 108 - 1 CCGUCGACGGCUGUGUGUUUAUUUGCCAUU-UUUUCUAGCCGUGUAUUUGUG--UGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGGCCAUUUGGCCAU ..(((((((((((((.((......))))).-......))))))(((((...(--(((....))))....)))))...))))...(((....)))..(((((....))))). ( -30.41) >consensus CCGUCGACGGCUGUAU_UUUAUUUCCCAUU_UUUUCUAGCCGUGUAUUUGUG__UGCACAUGCAUUAUUAAUGCCUCUGACAAUUGCAACUGCAUUUGG_CAUUUGGCCAU ..(((((((((((.......................)))))))((((.(((....))).))))..............))))...(((....)))..(((((....))))). (-17.69 = -18.97 + 1.28)

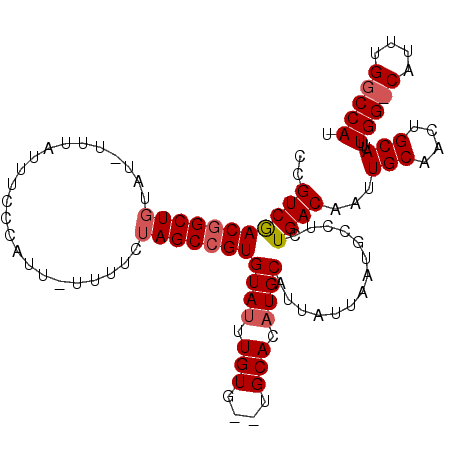
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:34 2006