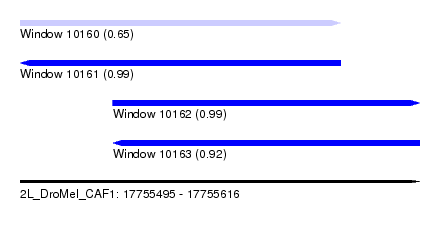
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,755,495 – 17,755,616 |
| Length | 121 |
| Max. P | 0.992299 |

| Location | 17,755,495 – 17,755,592 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.82 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -12.09 |
| Energy contribution | -13.95 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17755495 97 + 22407834 CUUCUAGUGUUGACUAAGGCCAUUU-----AAAGACUGCGUGUAAA---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUUAAGCCCUAG-- ....(((((((((...((((.....-----.......((((.....---------....))))(((((((((.......)))))))))..)))))))))))))........-- ( -25.60) >DroSec_CAF1 55469 97 + 1 UUUCUAGUGUUGACUAAGGCCAUUU-----AAAGACUGCGUGUAAA---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCUACAUAAAGCCCUAG-- .............(((.(((.....-----..(((..(((.(((..---------.....)))(((((((((.......))))))))).)))..))).......))).)))-- ( -21.94) >DroSim_CAF1 59330 97 + 1 UUUCUAGUGUUGACUAAGGCCAUUU-----AAAAACUGCGUGUAAA---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUAAAGCCCUAG-- ......(((((((...((((.....-----.......((((.....---------....))))(((((((((.......)))))))))..)))))))))))..........-- ( -23.80) >DroEre_CAF1 64540 97 + 1 GUGCUAGUGUUGGCUAAGGCCAUCU-----AGAGACUGCGUGUAAG---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUAAAGCCUUCG-- ...((((...((((....)))).))-----)).((((.......))---------))....(.(((((((((.......))))))))).)(((((......))))).....-- ( -28.50) >DroYak_CAF1 57775 105 + 1 -UGCUAGUGUUGGCUAAGUCCAUUU-----GAAGACUGCGUUUAAGGAAAAUGCAUUAAAUGCAAAGCCAUGUUUUCCACAAAGCAUUUCGCUUUCAAAAUAAAGCCUUCG-- -.(((.....(((((.((((.....-----...))))((((((((.(......).))))))))..)))))((((((....(((((.....))))).)))))).))).....-- ( -22.80) >DroAna_CAF1 63325 89 + 1 ---------------GAACCCACUUUGCCGUAAGUCAAGCUGUAAG---------CCGGAGGUUGAGCCGUGUUUUCAUCACAGCUUUUCCCAUUUGACACUGCACCUUAGGU ---------------..(((.....(((.((..(((((((.....)---------)..(.((..((((.(((.......))).))))..)))..))))))).))).....))) ( -20.60) >consensus _UUCUAGUGUUGACUAAGGCCAUUU_____AAAGACUGCGUGUAAA_________UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUAAAGCCCUAG__ ......(((((((...((((................(((((..................)))))((((((((.......))))))))...)))))))))))............ (-12.09 = -13.95 + 1.87)

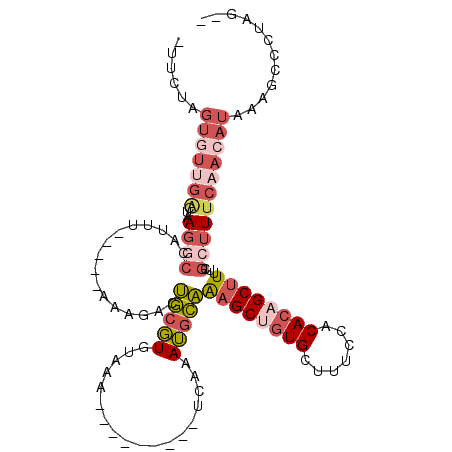
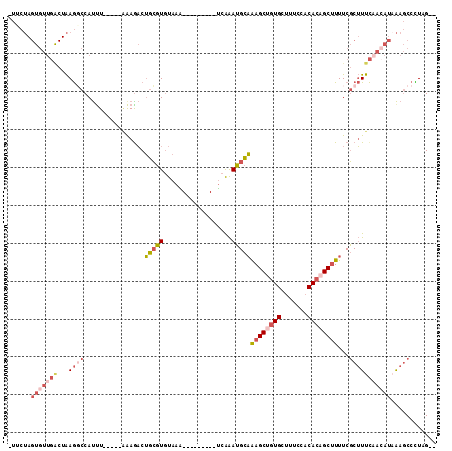
| Location | 17,755,495 – 17,755,592 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.82 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -14.13 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17755495 97 - 22407834 --CUAGGGCUUAAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------UUUACACGCAGUCUUU-----AAAUGGCCUUAGUCAACACUAGAAG --((((((((...((..((..(((.((((((((((.....)))))))))).....((.---------....)))))..))..)-----)...))))))))............. ( -29.90) >DroSec_CAF1 55469 97 - 1 --CUAGGGCUUUAUGUAGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------UUUACACGCAGUCUUU-----AAAUGGCCUUAGUCAACACUAGAAA --((((((((......(((..(((.((((((((((.....)))))))))).....((.---------....)))))..)))..-----....))))))))............. ( -31.10) >DroSim_CAF1 59330 97 - 1 --CUAGGGCUUUAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------UUUACACGCAGUUUUU-----AAAUGGCCUUAGUCAACACUAGAAA --((((((((.....((((((((..((((((((((.....))))))))))((......---------.......)).))))))-----))..))))))))............. ( -31.42) >DroEre_CAF1 64540 97 - 1 --CGAAGGCUUUAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------CUUACACGCAGUCUCU-----AGAUGGCCUUAGCCAACACUAGCAC --.((..((.....((..((.((((..((((((((.....)))))))))))).))..)---------)......))..)).((-----((.((((....))))...))))... ( -30.80) >DroYak_CAF1 57775 105 - 1 --CGAAGGCUUUAUUUUGAAAGCGAAAUGCUUUGUGGAAAACAUGGCUUUGCAUUUAAUGCAUUUUCCUUAAACGCAGUCUUC-----AAAUGGACUUAGCCAACACUAGCA- --.((((((......(((((.(((((..(((.(((.....))).)))))))).)))))(((.(((.....))).)))))))))-----...(((......))).........- ( -23.00) >DroAna_CAF1 63325 89 - 1 ACCUAAGGUGCAGUGUCAAAUGGGAAAAGCUGUGAUGAAAACACGGCUCAACCUCCGG---------CUUACAGCUUGACUUACGGCAAAGUGGGUUC--------------- (((((...(((.(((((((..(((...(((((((.(....))))))))...)))...(---------(.....)))))))..)).)))...)))))..--------------- ( -26.60) >consensus __CUAAGGCUUUAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA_________CUUACACGCAGUCUUU_____AAAUGGCCUUAGUCAACACUAGAA_ ....((((((.................((((((((.....))))))))((((......................))))..............))))))............... (-14.13 = -13.92 + -0.22)

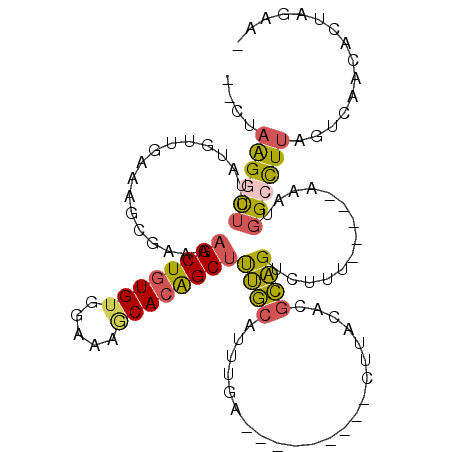
| Location | 17,755,523 – 17,755,616 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17755523 93 + 22407834 GACUGCGUGUAAA---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUUAAGCCCUAG-------GCAUUUUCGCUUCACUGUCAUCCA (((...(((....---------..(((((((((((((((.......)))))))))..((((........)))).....-------))))))......))).)))..... ( -25.00) >DroSec_CAF1 55497 93 + 1 GACUGCGUGUAAA---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCUACAUAAAGCCCUAG-------GCAUUUUCGCUUCACUGUCAUCCA (((...(((....---------..(((((((((((((((.......)))))))))..(((((......))))).....-------))))))......))).)))..... ( -25.30) >DroSim_CAF1 59358 93 + 1 AACUGCGUGUAAA---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUAAAGCCCUAG-------GCAUUUUCGCUUCACUGUCAUCCA ......(((....---------..(((((((((((((((.......)))))))))..(((((......))))).....-------))))))......)))......... ( -22.50) >DroEre_CAF1 64568 93 + 1 GACUGCGUGUAAG---------UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUAAAGCCUUCG-------GCAUUUCCACUUCACUGUCAUCCA (((...(((.(((---------(.(((((((((((((((.......)))))))))..(((((......))))).....-------))))))..))))))).)))..... ( -27.80) >DroYak_CAF1 57802 102 + 1 GACUGCGUUUAAGGAAAAUGCAUUAAAUGCAAAGCCAUGUUUUCCACAAAGCAUUUCGCUUUCAAAAUAAAGCCUUCG-------GCAUUUUCACUUCACUGUCAUCCA ((((((((((((.(......).)))))))))..((((((((((....)))))))...(((((......)))))....)-------))..............)))..... ( -21.90) >DroAna_CAF1 63343 100 + 1 GUCAAGCUGUAAG---------CCGGAGGUUGAGCCGUGUUUUCAUCACAGCUUUUCCCAUUUGACACUGCACCUUAGGUCCCACGUAUUCCUCUGUCCCUGUCAACCA .....((.....)---------).((.((..((((.(((.......))).))))..))...((((((..(((....(((...........))).)))...)))))))). ( -19.50) >consensus GACUGCGUGUAAA_________UCAAAUGCAAAGCUGUGCUUUCCACACAGCUUUUCGCUUUCAACAUAAAGCCCUAG_______GCAUUUUCGCUUCACUGUCAUCCA ........................(((((((((((((((.......)))))))))..((((........))))............)))))).................. (-13.32 = -13.85 + 0.53)
| Location | 17,755,523 – 17,755,616 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17755523 93 - 22407834 UGGAUGACAGUGAAGCGAAAAUGC-------CUAGGGCUUAAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------UUUACACGCAGUC .........(((((....((((((-------.....((((........))))..((((((((((.....))))))))))))))))..---------)))))........ ( -26.70) >DroSec_CAF1 55497 93 - 1 UGGAUGACAGUGAAGCGAAAAUGC-------CUAGGGCUUUAUGUAGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------UUUACACGCAGUC .........(((((....((((((-------.....(((((......)))))..((((((((((.....))))))))))))))))..---------)))))........ ( -27.90) >DroSim_CAF1 59358 93 - 1 UGGAUGACAGUGAAGCGAAAAUGC-------CUAGGGCUUUAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------UUUACACGCAGUU .........(((((....((((((-------.....(((((......)))))..((((((((((.....))))))))))))))))..---------)))))........ ( -27.90) >DroEre_CAF1 64568 93 - 1 UGGAUGACAGUGAAGUGGAAAUGC-------CGAAGGCUUUAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA---------CUUACACGCAGUC .....(((.(((((((.(((.(((-------.....(((((......)))))..((((((((((.....)))))))))))))))).)---------))).)))...))) ( -30.90) >DroYak_CAF1 57802 102 - 1 UGGAUGACAGUGAAGUGAAAAUGC-------CGAAGGCUUUAUUUUGAAAGCGAAAUGCUUUGUGGAAAACAUGGCUUUGCAUUUAAUGCAUUUUCCUUAAACGCAGUC .....(((.((((((.((((((((-------.....(((((......))))).((((((.(..((.....))..)....))))))...)))))))))))...))).))) ( -25.40) >DroAna_CAF1 63343 100 - 1 UGGUUGACAGGGACAGAGGAAUACGUGGGACCUAAGGUGCAGUGUCAAAUGGGAAAAGCUGUGAUGAAAACACGGCUCAACCUCCGG---------CUUACAGCUUGAC .((((((......((...((.(((...(.(((...))).).)))))...))......((((((.(....)))))))))))))((.((---------(.....))).)). ( -25.80) >consensus UGGAUGACAGUGAAGCGAAAAUGC_______CUAAGGCUUUAUGUUGAAAGCGAAAAGCUGUGUGGAAAGCACAGCUUUGCAUUUGA_________CUUACACGCAGUC ..............(((...................(((((......)))))(.((((((((((.....)))))))))).).....................))).... (-16.88 = -17.08 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:12 2006