
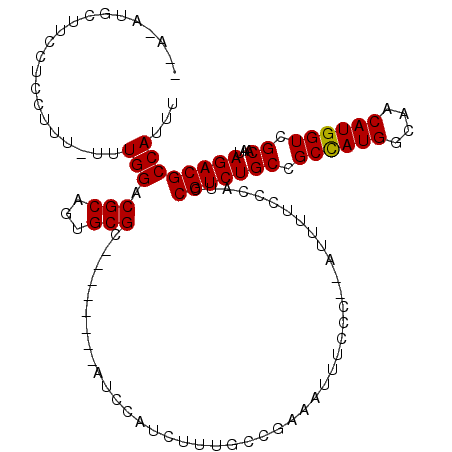
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,728,689 – 17,728,830 |
| Length | 141 |
| Max. P | 0.986083 |

| Location | 17,728,689 – 17,728,795 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -21.94 |
| Energy contribution | -21.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
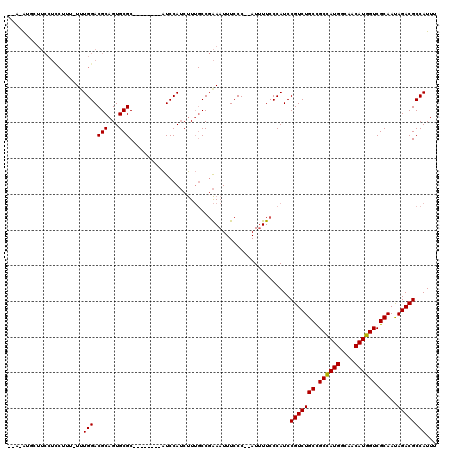
>2L_DroMel_CAF1 17728689 106 + 22407834 --A-AUGCUUCCUUCUUU-UUUGGACGCAGUGCGC--------AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUU --.-.(((.(((......-...))).)))(.((((--------(........))).((((......--..))))......((((((.((((((....)))))).))...))))))).... ( -28.70) >DroSec_CAF1 28909 107 + 1 --A-AUGCUUCCUCCUUUUUUUGGACGCAGUGCGC--------AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUU --.-.(((.(((..........))).)))(.((((--------(........))).((((......--..))))......((((((.((((((....)))))).))...))))))).... ( -28.60) >DroSim_CAF1 30267 107 + 1 --A-AUGCUUCCUCCUUUUUUUGGACGCAGUGCGC--------AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUU --.-.(((.(((..........))).)))(.((((--------(........))).((((......--..))))......((((((.((((((....)))))).))...))))))).... ( -28.60) >DroEre_CAF1 38706 109 + 1 UGAUAUUCUUCCUCCUUU-UUUGGACGCAGUGCGC--------AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAACAGACGCCAUUU .(((..............-..((((.((.....))--------.))))........((((......--..))))..)))((((((.(((((((....))))).))...))))))...... ( -26.40) >DroYak_CAF1 31130 107 + 1 --AUAGUCUUCCUCCUUU-UCUGGACGCAGUGCGC--------AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUU --...((((.........-...(((((..(((.((--------(........))).((((......--..)))).))).)))))((.((((((....)))))).))...))))....... ( -27.20) >DroAna_CAF1 38379 104 + 1 --A-GA----------UU-UUUGGACGCAGUGCGCCAGCCUCGGUCCAACCU--CCCGAAUUUCCCGAAUGUUCCCAUCCGUCUGCUGCUAUGGCAACAUGGUCGCAAUAGACGCCAUUU --.-..----------..-.((((((.(((.((....)))).)))))))...--...((((.........)))).....(((((((.((((((....)))))).))...)))))...... ( -27.20) >consensus __A_AUGCUUCCUCCUUU_UUUGGACGCAGUGCGC________AUCCAUCUUUGCCGAAAUUUCCC__AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUU .....................(((.(((...))).............................................(((((((.((((((....)))))).))...))))))))... (-21.94 = -21.80 + -0.14)

| Location | 17,728,720 – 17,728,830 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -25.67 |
| Energy contribution | -25.45 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
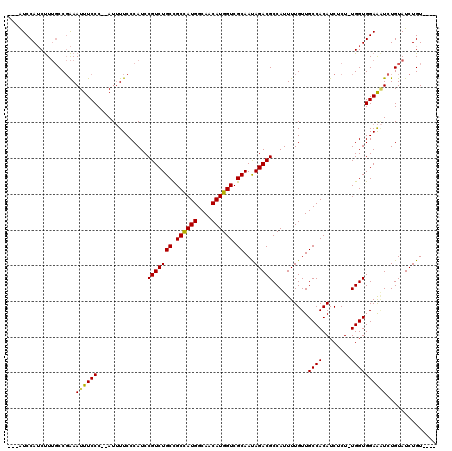
>2L_DroMel_CAF1 17728720 110 + 22407834 ---AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUUUGUUGCCACAUCUCU-UGGUGGAAAUCUGUAUCUGU---- ---.((((((......((((......--..)))).....(((((((.((((((....)))))).))...))))).....................-.)))))).............---- ( -27.50) >DroSec_CAF1 28941 110 + 1 ---AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUUUGUUGCCACAUCUCU-UGGUGGAAAUCUGUAUCUGU---- ---.((((((......((((......--..)))).....(((((((.((((((....)))))).))...))))).....................-.)))))).............---- ( -27.50) >DroSim_CAF1 30299 110 + 1 ---AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUUUGUUGCCACAUCUCU-UGGUGGAAAUCUGUAUCUGU---- ---.((((((......((((......--..)))).....(((((((.((((((....)))))).))...))))).....................-.)))))).............---- ( -27.50) >DroEre_CAF1 38740 110 + 1 ---AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAACAGACGCCAUUUUGUUGCCACAUCUCU-UGGUGGAAAUCUGUAUCUGU---- ---.((((((......((((......--..)))).....((((((.(((((((....))))).))...)))))).....................-.)))))).............---- ( -27.80) >DroYak_CAF1 31162 114 + 1 ---AUCCAUCUUUGCCGAAAUUUCCC--AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUUUGUUGCCACAUCUCU-UGGUGGAAUUCUGUAUCUGUAUUC ---.((((((......((((......--..)))).....(((((((.((((((....)))))).))...))))).....................-.))))))................. ( -27.50) >DroAna_CAF1 38405 114 + 1 UCGGUCCAACCU--CCCGAAUUUCCCGAAUGUUCCCAUCCGUCUGCUGCUAUGGCAACAUGGUCGCAAUAGACGCCAUUUUGUUGCCACAUCUCUUUGGUGGAGGUCUGUAUCUGU---- ..(((.((((((--((.......................(((((((.((((((....)))))).))...)))))..........((((........)))))))))).)).)))...---- ( -31.40) >consensus ___AUCCAUCUUUGCCGAAAUUUCCC__AUUUUCCCAUCCGUCUGCCGCCAUGGCAACAUGGUCGCAAUAGACGCCAUUUUGUUGCCACAUCUCU_UGGUGGAAAUCUGUAUCUGU____ ...................((((((..............(((((((.((((((....)))))).))...)))))..........((((........)))))))))).............. (-25.67 = -25.45 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:57 2006