
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,710,353 – 17,710,456 |
| Length | 103 |
| Max. P | 0.972493 |

| Location | 17,710,353 – 17,710,456 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -24.44 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.00 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17710353 103 + 22407834 UACUUCGACAGUACGCGAUAAUGGCGGAUAGCCCUUGUCUUGGGCCAUGGCCUCUAACUUGUAUGUCUCGCGCUGUAU----------------UUUAUAUACAUUUCACGAUCAAUAU- ......((((.(((..(((((.(((.....))).)))))..((((....)))).......)))))))(((.(.(((((----------------.....)))))...).))).......- ( -25.30) >DroVir_CAF1 8475 95 + 1 UACUUCGACCGUACGCGAUAAUGGCGGAUAGCCCUUGUCUUGGGCCAUCGCCUCUAAUUUGUAUGUCUCGCGCUGUUU----------------UUUGAAUACAUAUUCAU--------- .....(((.((((((.......((((.((.((((.......)))).)))))).......))))))..)))........----------------..(((((....))))).--------- ( -22.94) >DroGri_CAF1 11871 104 + 1 UACUUCGACCGUUCGCGAUAAUGGCGGAUAGCCCUUGUCUUGGGCCAUCGCCUCUAAUUUGUAUGUCUCGCGCUGUAU----------------UUCAAAUACAUUUUCAAUUCAAUUUG .............(((((((..((((.((.((((.......)))).))))))..))...........))))).(((((----------------.....)))))................ ( -20.80) >DroWil_CAF1 7115 115 + 1 UACCUCGACAGUACGAGAUAAUGGCGGAUAGCCUUUGUCUUGGGCCAUUGCCUCUAAUUUGUAUGUCUCGCGCUGUAUAUAUCAUUUGCGGUUUAUUAUAUACAUUUU----UUUUUGU- .......((((..((((((((.(((.....))).))))))))(((....))).......(((((((.....((((((.........)))))).....)))))))....----...))))- ( -27.40) >DroMoj_CAF1 9488 92 + 1 CACUUCGACCGUGCGCGAUAAUGGCGGAUAGCCCUUGUCUUGGGCCAUCGCCUCUAAUUUGUAUGUCUCGCGCUGUAU----------------UUUGAAUGCGA---UAG--------- ..........(((((.((((..((((.((.((((.......)))).)))))).(......)..)))).)))))(((((----------------.....))))).---...--------- ( -25.40) >DroPer_CAF1 38976 91 + 1 UACUUCGACAGUACGCGAUAGUGGCGGAUAGCCCUUGUCUUGGGCCAUGGCCUCUAACUUGUAUGUCUCGCGCUGUAU----------------UUUAUAUACAUUU------------- ......((((.((((.(.(((.(((.(...((((.......))))..).))).))).).))))))))......(((((----------------.....)))))...------------- ( -24.80) >consensus UACUUCGACAGUACGCGAUAAUGGCGGAUAGCCCUUGUCUUGGGCCAUCGCCUCUAAUUUGUAUGUCUCGCGCUGUAU________________UUUAAAUACAUUUU_A__________ ......(((.((((..(((((.(((.....))).)))))..((((....)))).......)))))))..................................................... (-19.08 = -19.00 + -0.08)

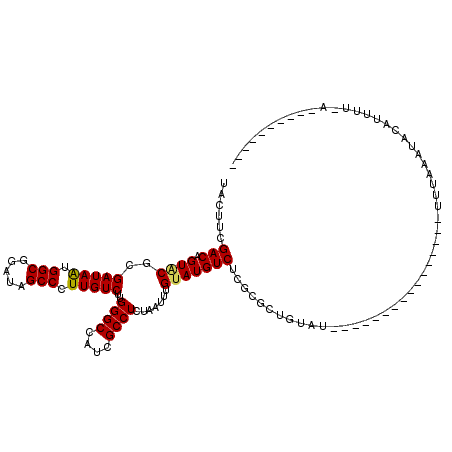

| Location | 17,710,353 – 17,710,456 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -19.43 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17710353 103 - 22407834 -AUAUUGAUCGUGAAAUGUAUAUAAA----------------AUACAGCGCGAGACAUACAAGUUAGAGGCCAUGGCCCAAGACAAGGGCUAUCCGCCAUUAUCGCGUACUGUCGAAGUA -.......(((((...(((((.....----------------))))).)))))(((((((..(.(((.(((.(((((((.......)))))))..))).))).)..))).))))...... ( -31.50) >DroVir_CAF1 8475 95 - 1 ---------AUGAAUAUGUAUUCAAA----------------AAACAGCGCGAGACAUACAAAUUAGAGGCGAUGGCCCAAGACAAGGGCUAUCCGCCAUUAUCGCGUACGGUCGAAGUA ---------.(((((....)))))..----------------.....((((((......(......).(((((((((((.......))))))).))))....))))))............ ( -28.70) >DroGri_CAF1 11871 104 - 1 CAAAUUGAAUUGAAAAUGUAUUUGAA----------------AUACAGCGCGAGACAUACAAAUUAGAGGCGAUGGCCCAAGACAAGGGCUAUCCGCCAUUAUCGCGAACGGUCGAAGUA ....((((.(((....(((((.....----------------))))).(((((......(......).(((((((((((.......))))))).))))....)))))..))))))).... ( -29.20) >DroWil_CAF1 7115 115 - 1 -ACAAAAA----AAAAUGUAUAUAAUAAACCGCAAAUGAUAUAUACAGCGCGAGACAUACAAAUUAGAGGCAAUGGCCCAAGACAAAGGCUAUCCGCCAUUAUCUCGUACUGUCGAGGUA -.......----....((((((((...............))))))))((....).)............(((.((((((.........))))))..)))..(((((((......))))))) ( -23.46) >DroMoj_CAF1 9488 92 - 1 ---------CUA---UCGCAUUCAAA----------------AUACAGCGCGAGACAUACAAAUUAGAGGCGAUGGCCCAAGACAAGGGCUAUCCGCCAUUAUCGCGCACGGUCGAAGUG ---------...---.(((.(((...----------------((...((((((......(......).(((((((((((.......))))))).))))....))))))...)).)))))) ( -29.10) >DroPer_CAF1 38976 91 - 1 -------------AAAUGUAUAUAAA----------------AUACAGCGCGAGACAUACAAGUUAGAGGCCAUGGCCCAAGACAAGGGCUAUCCGCCACUAUCGCGUACUGUCGAAGUA -------------...(((((.....----------------)))))(((((((((......)))((.(((.(((((((.......)))))))..))).)).))))))(((.....))). ( -27.70) >consensus __________U_AAAAUGUAUAUAAA________________AUACAGCGCGAGACAUACAAAUUAGAGGCGAUGGCCCAAGACAAGGGCUAUCCGCCAUUAUCGCGUACGGUCGAAGUA ...............................................((((((......(......).(((.(((((((.......)))))))..)))....))))))............ (-19.43 = -19.82 + 0.39)

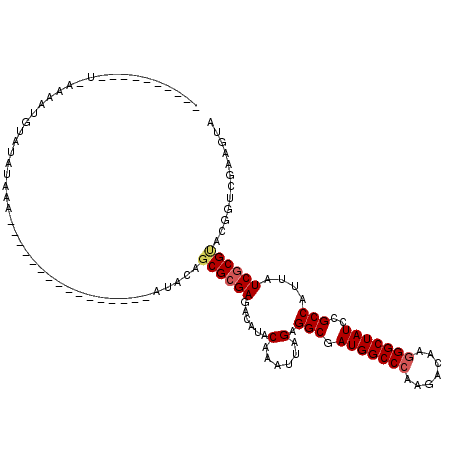

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:48 2006