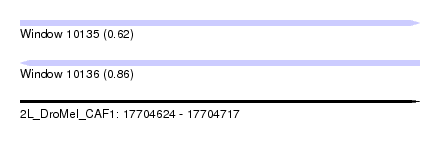
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,704,624 – 17,704,717 |
| Length | 93 |
| Max. P | 0.858693 |

| Location | 17,704,624 – 17,704,717 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -13.05 |
| Energy contribution | -12.85 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17704624 93 + 22407834 UUCUGCAUUCUGCAUUAGUUUUUUUC-AC-AACGCCCUCCUUA--UUCGUGGACUUUC--ACCAAGGGCUUAGCACACACUUGGAAGGCGUGCAAAAAU ...(((.....)))......(((((.-..-.(((((.(((...--...(((..((...--.(....)....))..)))....))).)))))..))))). ( -18.90) >DroPse_CAF1 32072 95 + 1 UUCUGCAUUUUGCAUUAGUUU-UUUU-UUGUUUUGCCUCUCGC--UUGCUGGCCCUUCCAUCCGAGGGCUUAGCACACACUUGGAAGGCGUGCAAAAAU ...(((.....))).......-.(((-((((..(((((..((.--.(((((((((((......)))))).)))))......))..))))).))))))). ( -29.70) >DroEre_CAF1 16530 92 + 1 CCCUGCAUUUUGCAUUAGUUU-UUUC-AA-AACGCCCUCCUUA--GUCCUGGACUUUC--ACCAAGGGCUUAGCACACACUUGGAAGGCGUGCAAAAAU .......((((((((..((((-....-.)-)))(((.(((..(--(((((((......--.)).))))))............))).))))))))))).. ( -24.64) >DroYak_CAF1 8499 92 + 1 UUCUGCAUUUUGCAUUAGUUU-UUUC-AA-AACGCCCUCCUUA--GUCCUGGACUUUC--ACCAAGGGCUUAGCACACACUUGGAAGGCGUGCAAGAAU .......((((((((..((((-....-.)-)))(((.(((..(--(((((((......--.)).))))))............))).))))))))))).. ( -24.74) >DroAna_CAF1 16821 88 + 1 CUCUGCAUUUUGCAUUAGUUU-CC--------CGCCCUGUCUCCCACUUUGGACUUUC--ACCAAGGGCUUAGCACACACUUGGAAGGCGUGCAAAAAU .......((((((((...(((-((--------.....(((((....((((((......--.))))))....)).))).....)))))..)))))))).. ( -19.90) >DroPer_CAF1 32589 96 + 1 UUCUGCAUUUUGCAUUAGUUU-UUUUUUUUUUUUGCCUCUCGC--CGGCUGGCCCUUCCAUCCGAGGGCUUAGCACACACUUGGAAGGCGUGCAAAAAU ...(((.....))).......-.......(((((((....(((--(.((((((((((......)))))).)))).((....))...)))).))))))). ( -28.20) >consensus UUCUGCAUUUUGCAUUAGUUU_UUUC_AA_AACGCCCUCCCUA__GUCCUGGACUUUC__ACCAAGGGCUUAGCACACACUUGGAAGGCGUGCAAAAAU .......((((((((..((((..............((.............)).........(((((.((...)).....))))).)))))))))))).. (-13.05 = -12.85 + -0.19)

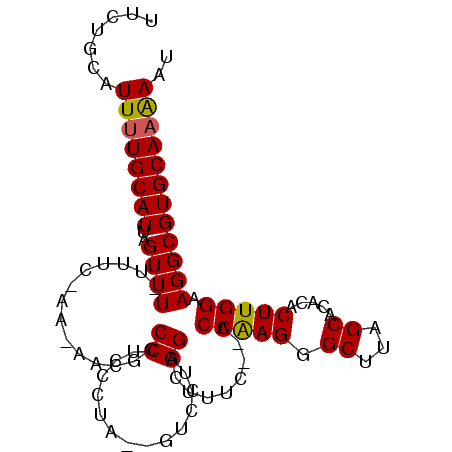
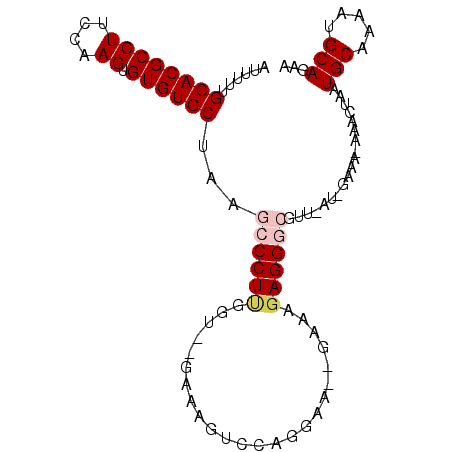
| Location | 17,704,624 – 17,704,717 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -16.59 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17704624 93 - 22407834 AUUUUUGCACGCCUUCCAAGUGUGUGCUAAGCCCUUGGU--GAAAGUCCACGAA--UAAGGAGGGCGUU-GU-GAAAAAAACUAAUGCAGAAUGCAGAA .((((..((((((((((...((((..((..(((...)))--...))..))))..--...)))))))).)-).-.)))).......(((.....)))... ( -31.80) >DroPse_CAF1 32072 95 - 1 AUUUUUGCACGCCUUCCAAGUGUGUGCUAAGCCCUCGGAUGGAAGGGCCAGCAA--GCGAGAGGCAAAACAA-AAAA-AAACUAAUGCAAAAUGCAGAA .((((((...(((((....((...((((..(((((........))))).)))).--))..)))))....)))-))).-.......(((.....)))... ( -25.20) >DroEre_CAF1 16530 92 - 1 AUUUUUGCACGCCUUCCAAGUGUGUGCUAAGCCCUUGGU--GAAAGUCCAGGAC--UAAGGAGGGCGUU-UU-GAAA-AAACUAAUGCAAAAUGCAGGG ..(((((((((((((((.(((....))).....((((((--....).)))))..--...)))))))))(-((-....-))).....))))))....... ( -27.40) >DroYak_CAF1 8499 92 - 1 AUUCUUGCACGCCUUCCAAGUGUGUGCUAAGCCCUUGGU--GAAAGUCCAGGAC--UAAGGAGGGCGUU-UU-GAAA-AAACUAAUGCAAAAUGCAGAA .(((....(((((((((.(((....))).....((((((--....).)))))..--...))))))))).-..-))).-.......(((.....)))... ( -26.10) >DroAna_CAF1 16821 88 - 1 AUUUUUGCACGCCUUCCAAGUGUGUGCUAAGCCCUUGGU--GAAAGUCCAAAGUGGGAGACAGGGCG--------GG-AAACUAAUGCAAAAUGCAGAG ......((((((((....)).))))))...(((((((((--....).))))....(....).))))(--------(.-...))..(((.....)))... ( -26.60) >DroPer_CAF1 32589 96 - 1 AUUUUUGCACGCCUUCCAAGUGUGUGCUAAGCCCUCGGAUGGAAGGGCCAGCCG--GCGAGAGGCAAAAAAAAAAAA-AAACUAAUGCAAAAUGCAGAA .(((((((...(((((((..((.(.((...)).).))..)))))))(((....)--)).....))))))).......-.......(((.....)))... ( -25.80) >consensus AUUUUUGCACGCCUUCCAAGUGUGUGCUAAGCCCUUGGU__GAAAGUCCAGGAA__GAAAGAGGGCGUU_AU_GAAA_AAACUAAUGCAAAAUGCAGAA ......((((((((....)).))))))...((((((........................))))))...................(((.....)))... (-16.59 = -17.26 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:45 2006