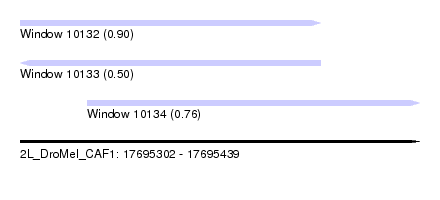
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 17,695,302 – 17,695,439 |
| Length | 137 |
| Max. P | 0.899984 |

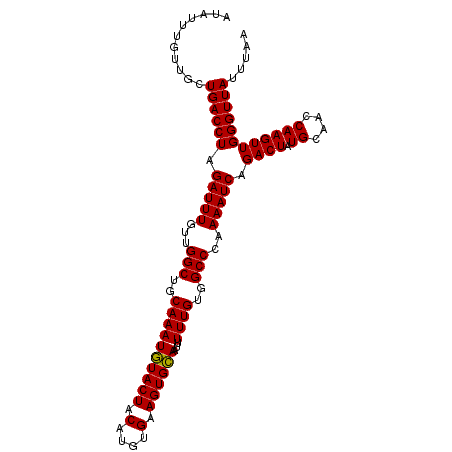
| Location | 17,695,302 – 17,695,405 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 98.54 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.22 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17695302 103 + 22407834 AUAUUUGUUGCUGACCUAGAUUUGUUGGCUGCAAAUUUACUACAUGUGAAGUGAAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAA ...........((((((.(((((...(((..((((((((((.(....).))))))...))))..)))..))))).((((.((....))))))))))))..... ( -25.00) >DroSec_CAF1 86107 103 + 1 AUAUUUGUUGCUGACCUAGAUUUGUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAA ...........((((((.(((((...(((..((((((((((.(....).))))))...))))..)))..))))).((((.((....))))))))))))..... ( -27.40) >DroSim_CAF1 68667 103 + 1 AUAUUUGUUGCUGACCUAGAUUUGUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAA ...........((((((.(((((...(((..((((((((((.(....).))))))...))))..)))..))))).((((.((....))))))))))))..... ( -27.40) >DroYak_CAF1 89898 103 + 1 AUAUUUGUUGCUGACCUAGAUUUGUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAAGCAAGUUGGGUUAUUUAA ...........((((((.(((((...(((..((((((((((.(....).))))))...))))..)))..))))).((((.((....))))))))))))..... ( -29.10) >consensus AUAUUUGUUGCUGACCUAGAUUUGUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAA ...........((((((.(((((...(((..((((((((((.(....).))))))...))))..)))..))))).((((.((....))))))))))))..... (-27.60 = -27.22 + -0.38)

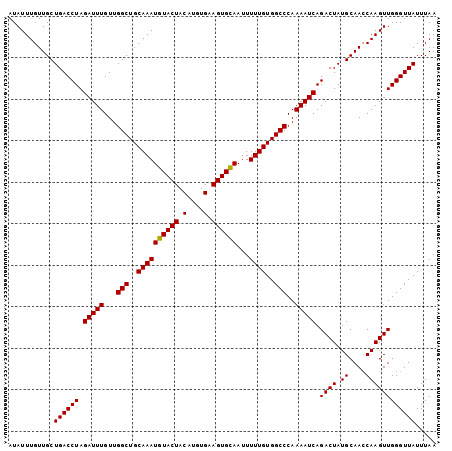
| Location | 17,695,302 – 17,695,405 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 98.54 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17695302 103 - 22407834 UUAAAUAACCCAACUUGGUUGCAUAGUCUGAUUUUGGGCCACAAAAAUUUCACUUCACAUGUAGUAAAUUUGCAGCCAACAAAUCUAGGUCAGCAACAAAUAU .................(((((.(..((((((((..(((.....((((((.(((........)))))))))...)))...))))).)))..))))))...... ( -19.10) >DroSec_CAF1 86107 103 - 1 UUAAAUAACCCAACUUGGUUGCAUAGUCUGAUUUUGGGCCACAAAAAUUGCACUUCACAUGUAGUACAUUUGCAGCCAACAAAUCUAGGUCAGCAACAAAUAU .................(((((.(..((((((((..(((..((((.((((((.......))))))...))))..)))...))))).)))..))))))...... ( -20.90) >DroSim_CAF1 68667 103 - 1 UUAAAUAACCCAACUUGGUUGCAUAGUCUGAUUUUGGGCCACAAAAAUUGCACUUCACAUGUAGUACAUUUGCAGCCAACAAAUCUAGGUCAGCAACAAAUAU .................(((((.(..((((((((..(((..((((.((((((.......))))))...))))..)))...))))).)))..))))))...... ( -20.90) >DroYak_CAF1 89898 103 - 1 UUAAAUAACCCAACUUGCUUGCAUAGUCUGAUUUUGGGCCACAAAAAUUGCACUUCACAUGUAGUACAUUUGCAGCCAACAAAUCUAGGUCAGCAACAAAUAU ..............(((.((((.(..((((((((..(((..((((.((((((.......))))))...))))..)))...))))).)))..)))))))).... ( -18.80) >consensus UUAAAUAACCCAACUUGGUUGCAUAGUCUGAUUUUGGGCCACAAAAAUUGCACUUCACAUGUAGUACAUUUGCAGCCAACAAAUCUAGGUCAGCAACAAAUAU .................(((((.(..((((((((..(((..((((.((((((.......))))))...))))..)))...))))).)))..))))))...... (-18.42 = -18.92 + 0.50)

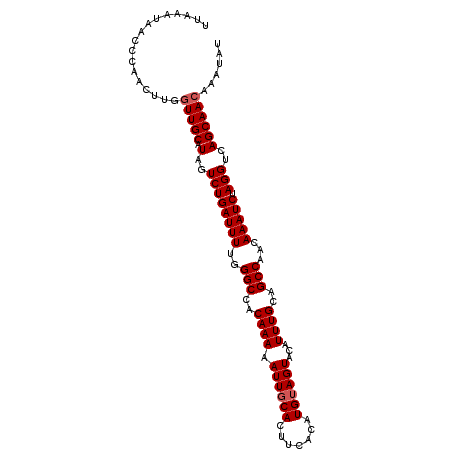
| Location | 17,695,325 – 17,695,439 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.05 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -27.99 |
| Energy contribution | -28.68 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 17695325 114 + 22407834 GUUGGCUGCAAAUUUACUACAUGUGAAGUGAAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAAAUCGCAAACAUGUGUACGGGUAACCGCUCGCCCA ...(((.((.....(((.((((((...((((..(((..(((((((((....................)))))))))..)))))))..))))))))).((....))))..))).. ( -29.75) >DroSec_CAF1 86130 114 + 1 GUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAAAUCGCAAACAAGUGUACGGGUAACCGCUCGACAA ((((((.......((((.((.(((....(((.((((..(((((((((....................)))))))))..)))).))).))).))))))((....)))).)))).. ( -31.35) >DroSim_CAF1 68690 114 + 1 GUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAAAUCGCAAACAUGUGUACGGGUAACCGCUCGCCCA ...(((.((....((((.((((((....(((.((((..(((((((((....................)))))))))..)))).))).))))))))))((....))))..))).. ( -35.85) >DroYak_CAF1 89921 114 + 1 GUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAAGCAAGUUGGGUUAUUUAAAUCGCAAACAUGUGUAUGGCUAACCACUCGCCCA (((((((......((((.((((((....(((.((((..((((((((.......(((.((....)))))))))))))..)))).))).))))))))))))))))).......... ( -32.51) >consensus GUUGGCUGCAAAUGUACUACAUGUGAAGUGCAAUUUUUGUGGCCCAAAAUCAGACUAUGCAACCAAGUUGGGUUAUUUAAAUCGCAAACAUGUGUACGGGUAACCGCUCGCCCA ..(((.(((...(((((.((((((....(((.((((..(((((((((....................)))))))))..)))).))).))))))))))).))).)))........ (-27.99 = -28.68 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:43 2006